|
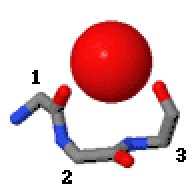
Niche (protein Structural Motif)
In the area of protein Structural motif, structural motifs, niches are three or four amino acid residue features in which main-chain CO groups are bridged by positively charged or δ+ groups. The δ+ groups include groups with two hydrogen bond donor atoms such as NH2 groups and water molecules. In typical proteins, 7% of amino acid residues belong to niches bound to a δ+ group, while another 7% have the conformation but no single Cation, cationic bridging group is detected. Niches are of two kinds, distinguished as niche3 (3 residues, ''i'' to ''i+2'') and niche4 (4 residues, ''i'' to ''i+3''). In a niche3 motif the δ+-binding carbonyl groups are from residues ''i'' and ''i+2'' while in a niche4 motif they are from residues ''i'' and ''i+3''. A niche3 has the α conformation for residue ''i+1'' and the β conformation for residue ''i+2''; a niche4 has the α conformation for residues ''i+1'' and ''i+2'' and the β conformation for residue ''i+3''. A niche occurs commonly at the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metabolic reactions, DNA replication, Cell signaling, responding to stimuli, providing Cytoskeleton, structure to cells and Fibrous protein, organisms, and Intracellular transport, transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the Nucleic acid sequence, nucleotide sequence of their genes, and which usually results in protein folding into a specific Protein structure, 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called pep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Annexin
Annexin is a common name for a group of cellular proteins. They are mostly found in eukaryotic organisms (animals, plants and fungi). In humans, the annexins are found inside the cell. However some annexins (Annexin A1, Annexin A2, and Annexin A5) can be secreted from the cytoplasm to outside cellular environments, such as blood. Annexin is also known as ''lipocortin''. Lipocortins suppress phospholipase A2. Increased expression of the gene coding for annexin-1 is one of the mechanisms by which glucocorticoids (such as cortisol) inhibit inflammation. Introduction The protein family of annexins has continued to grow since their association with intracellular membranes was first reported in 1977. The recognition that these proteins were members of a broad family first came from protein sequence comparisons and their cross-reactivity with antibodies. One of these workers (Geisow) coined the name Annexin shortly after. As of 2002 160 annexin proteins have been identified in 65 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hsp70
The 70 kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms and play crucial roles in the development of cancer, neurodegeneration, apoptosis, regulating sleep, and much more. Intracellularly localized Hsp70s are an important part of the cell's machinery for protein folding, performing chaperoning functions, and helping to protect cells from the adverse effects of physiological stresses. Additionally, membrane-bound Hsp70s have been identified as a potential target for cancer therapies and their extracellularly localized counterparts have been identified as having both membrane-bound and membrane-free structures. There is lot of potential in the Hsp70 protein as a key therapeutic target for developing new drugs for the treatment of sleep disorders, cancer, neurodegeneration, and other related pathological conditions. Discovery Members of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Export Signal
A nuclear export signal (NES) is a short target peptide containing 4 hydrophobic residues in a protein that targets it for export from the cell nucleus to the cytoplasm through the nuclear pore complex using nuclear transport. It has the opposite effect of a nuclear localization signal, which targets a protein located in the cytoplasm for import to the nucleus. The NES is recognized and bound by exportins. NESs serve several vital cellular functions. They assist in regulating the position of proteins within the cell. Through this NESs affect transcription and several other nuclear functions that are essential to proper cell function. The export of many types of RNA from the nucleus is required for proper cellular function. The NES determines what type of pathway the varying types of RNA may use to exit the nucleus and perform their function and the NESs may effect the directionality of molecules exiting the nucleus. Structure Computer analysis of known NESs found the most com ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group (which is in the deprotonated form when the lysine is dissolved in water at physiological pH), and a side chain (which is partially protonated when the lysine is dissolved in water at physiological pH), and so it is classified as a basic, charged (in water at physiological pH), aliphatic amino acid. It is encoded by the codons AAA and AAG. Like almost all other amino acids, the α-carbon is chiral and lysine may refer to either enantiomer or a racemic mixture of both. For the purpose of this article, lysine will refer to the biologically active enantiomer L-lysine, where the α-carbon is in the ''S'' configuration. The human body cannot synthesize lysine. It is essential in humans and must therefore be obtained from the diet. In orga ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carbonyl Group
In organic chemistry, a carbonyl group is a functional group with the formula , composed of a carbon atom double-bonded to an oxygen atom, and it is divalent at the C atom. It is common to several classes of organic compounds (such as aldehydes, ketones and carboxylic acid), as part of many larger functional groups. A compound containing a carbonyl group is often referred to as a carbonyl compound. The term carbonyl can also refer to carbon monoxide as a ligand in an inorganic or organometallic complex (a metal carbonyl, e.g. nickel carbonyl). The remainder of this article concerns itself with the organic chemistry definition of carbonyl, such that carbon and oxygen share a double bond. Carbonyl compounds In organic chemistry, a carbonyl group characterizes the following types of compounds: Other organic carbonyls are urea and the carbamates, the derivatives of acyl chlorides, chloroformates and phosgene, carbonate esters, thioesters, lactones, lactams, hydroxama ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Domain
A transmembrane domain (TMD, TM domain) is a membrane-spanning protein domain. TMDs may consist of one or several alpha-helices or a transmembrane beta barrel. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in TMDs are often hydrophobic, although proteins such as membrane pumps and ion channels can contain polar residues. TMDs vary greatly in size and hydrophobicity; they may adopt organelle-specific properties. Functions of transmembrane domains Transmembrane domains are known to perform a variety of functions. These include: * Anchoring transmembrane proteins to the membrane. *Facilitating molecular transport of molecules such as ions and proteins across biological membranes; usually hydrophilic residues and binding sites in the TMDs help in this process. *Signal transduction across the membrane; many transmembrane proteins, such as G protein-coupled receptors, receive extracellular signals. TMDs then propagate those signals across the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyruvate Dehydrogenase
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate. Pyruvate dehydrogenase is usually encountered as a component, referred to as E1, of the pyruvate dehydrogenase complex (PDC). PDC consists of other enzymes, referred to as E2 and E3. Collectively E1-E3 transform pyruvate, NAD+, coenzyme A into acetyl-CoA, CO2, and NADH. The conversion is crucial because acetyl-CoA may then be used in the citric acid cycle to carry out cellular respiration. To distinguish between this enzyme and the PDC, it is systematically called pyruvate dehydrogenase (acetyl-transferring). Mechanism The thiamine pyrophosphate (TPP) converts to an ylide by deprotonation. The ylide attack the ketone group of pyruvate. The resulting adduct decarboxylates. The resulting 1,3-dipole reductively acetylates lipoamide-E2. In terms of details, biochem ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thrombin
Prothrombin (coagulation factor II) is encoded in the human by the F2-gene. It is proteolytically cleaved during the clotting process by the prothrombinase enzyme complex to form thrombin. Thrombin (Factor IIa) (, fibrose, thrombase, thrombofort, topical, thrombin-C, tropostasin, activated blood-coagulation factor II, E thrombin, beta-thrombin, gamma-thrombin) is a serine protease, that converts fibrinogen into strands of insoluble fibrin, as well as catalyzing many other coagulation-related reactions. History After the description of fibrinogen and fibrin, Alexander Schmidt hypothesised the existence of an enzyme that converts fibrinogen into fibrin in 1872. Prothrombin was discovered by Pekelharing in 1894. Physiology Synthesis Thrombin is produced by the enzymatic cleavage of two sites on prothrombin by activated Factor X (Xa). The activity of factor Xa is greatly enhanced by binding to activated Factor V (Va), termed the prothrombinase complex. Prothrombin is prod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Motif
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA. In nucleic acids Depending upon the sequence and other conditions, nucleic acids can form a variety of structural motifs which is thought to have biological significance. ;Stem-loop: Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
310 Helix
A 310 helix is a type of secondary structure found in proteins and polypeptides. Of the numerous protein secondary structures present, the 310-helix is the fourth most common type observed; following α-helices, β-sheets and reverse turns. 310-helices constitute nearly 10–15% of all helices in protein secondary structures, and are typically observed as extensions of α-helices found at either their N- or C- termini. Because of the α-helices tendency to consistently fold and unfold, it has been proposed that the 310-helix serves as an intermediary conformation of sorts, and provides insight into the initiation of α-helix folding. Discovery Max Perutz, the head of the Medical Research Council Laboratory of Molecular Biology at the University of Cambridge, wrote the first paper documenting the elusive 310-helix. Together with Lawrence Bragg and John Kendrew, Perutz published an exploration of polypeptide chain configurations in 1950, based on cues from noncrystalline diff ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |