|
Glycogen Phosphorylase
Glycogen phosphorylase is one of the phosphorylase enzymes (). Glycogen phosphorylase catalyzes the rate-limiting step in glycogenolysis in animals by releasing glucose-1-phosphate from the terminal alpha-1,4-glycosidic bond. Glycogen phosphorylase is also studied as a model protein regulated by both reversible phosphorylation and allosteric effects. Mechanism Glycogen phosphorylase breaks up glycogen into glucose subunits (see also figure below): (α-1,4 glycogen chain)n + Pi ⇌ (α-1,4 glycogen chain)n-1 + α-D-glucose-1-phosphate. Glycogen is left with one fewer glucose molecule, and the free glucose molecule is in the form of glucose-1-phosphate. In order to be used for metabolism, it must be converted to glucose-6-phosphate by the enzyme phosphoglucomutase. Although the reaction is reversible in vitro, within the cell the enzyme only works in the forward direction as shown below because the concentration of inorganic phosphate is much higher than that of gluco ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphorylase
In biochemistry, phosphorylases are enzymes that catalyze the addition of a phosphate group from an inorganic phosphate (phosphate+hydrogen) to an acceptor. :A-B + P A + P-B They include allosteric enzymes that catalyze the production of glucose-1-phosphate from a glucan such as glycogen, starch or maltodextrin. Phosphorylase is also a common name used for glycogen phosphorylase in honor of Earl W. Sutherland Jr., who in the late 1930s discovered it as the first phosphorylase. Function Phosphorylases should not be confused with phosphatases, which remove phosphate groups. In more general terms, phosphorylases are enzymes that catalyze the addition of a phosphate group from an inorganic phosphate (phosphate + hydrogen) to an acceptor, not to be confused with a phosphatase (a hydrolase) or a kinase (a phosphotransferase). A phosphatase removes a phosphate group from a donor using water, whereas a kinase transfers a phosphate group from a donor (usually ATP) to an accep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Branching (polymer Chemistry)
In polymer chemistry, branching is the regular or irregular attachment of side chains to a polymer's backbone chain. It occurs by the replacement of a substituent (e.g. a hydrogen atom) on a monomer subunit by another covalently-bonded chain of that polymer; or, in the case of a graft copolymer, by a chain of another type. Branched polymers have more compact and symmetrical molecular conformations, and exhibit intra-heterogeneous dynamical behavior with respect to the unbranched polymers. In crosslinking rubber by vulcanization, short sulfur branches link polyisoprene chains (or a synthetic variant) into a multiple-branched thermosetting elastomer. Rubber can also be so completely vulcanized that it becomes a rigid solid, so hard it can be used as the bit in a smoking pipe. Polycarbonate chains can be crosslinked to form the hardest, most impact-resistant thermosetting plastic, used in safety glasses. Branching may result from the formation of carbon-carbon or vario ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allosteric
In the fields of biochemistry and pharmacology an allosteric regulator (or allosteric modulator) is a substance that binds to a site on an enzyme or receptor distinct from the active site, resulting in a conformational change that alters the protein's activity, either enhancing or inhibiting its function. In contrast, substances that bind directly to an enzyme's active site or the binding site of the endogenous ligand of a receptor are called orthosteric regulators or modulators. The site to which the effector binds is termed the ''allosteric site'' or ''regulatory site''. Allosteric sites allow effectors to bind to the protein, often resulting in a conformational change and/or a change in protein dynamics. Effectors that enhance the protein's activity are referred to as ''allosteric activators'', whereas those that decrease the protein's activity are called ''allosteric inhibitors''. Allosteric regulations are a natural example of control loops, such as feedback from downs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalytic
Catalysis () is the increase in reaction rate, rate of a chemical reaction due to an added substance known as a catalyst (). Catalysts are not consumed by the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quickly, very small amounts of catalyst often suffice; mixing, surface area, and temperature are important factors in reaction rate. Catalysts generally react with one or more reactants to form reaction intermediate, intermediates that subsequently give the final reaction product, in the process of regenerating the catalyst. The rate increase occurs because the catalyst allows the reaction to occur by an alternative mechanism which may be much faster than the noncatalyzed mechanism. However the noncatalyzed mechanism does remain possible, so that the total rate (catalyzed plus noncatalyzed) can only increase in the presence of the catalyst and never decrease. Catalysis may be classified as either homogeneous catalysis, homogeneou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isozyme
In biochemistry, isozymes (also known as isoenzymes or more generally as multiple forms of enzymes) are enzymes that differ in amino acid sequence but catalyze the same chemical reaction. Isozymes usually have different kinetic parameters (e.g. different ''K''M values), or are regulated differently. They permit the fine-tuning of metabolism to meet the particular needs of a given tissue or developmental stage. In many cases, isozymes are encoded by homologous genes that have diverged over time. Strictly speaking, enzymes with different amino acid sequences that catalyse the same reaction are isozymes if encoded by different genes, or allozymes if encoded by different alleles of the same gene; the two terms are often used interchangeably. Introduction Isozymes were first described by R. L. Hunter and Clement Markert (1957) who defined them as ''different variants of the same enzyme having identical functions and present in the same individual''. This definition encompasses ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dimer
In biochemistry, a protein dimer is a macromolecular complex or protein multimer, multimer formed by two protein monomers, or single proteins, which are usually Non-covalent interaction, non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", ''wikt:di-#Prefix, di-'' + ''wikt:-mer#Suffix, -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins while a protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein IKBKG, NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleophile
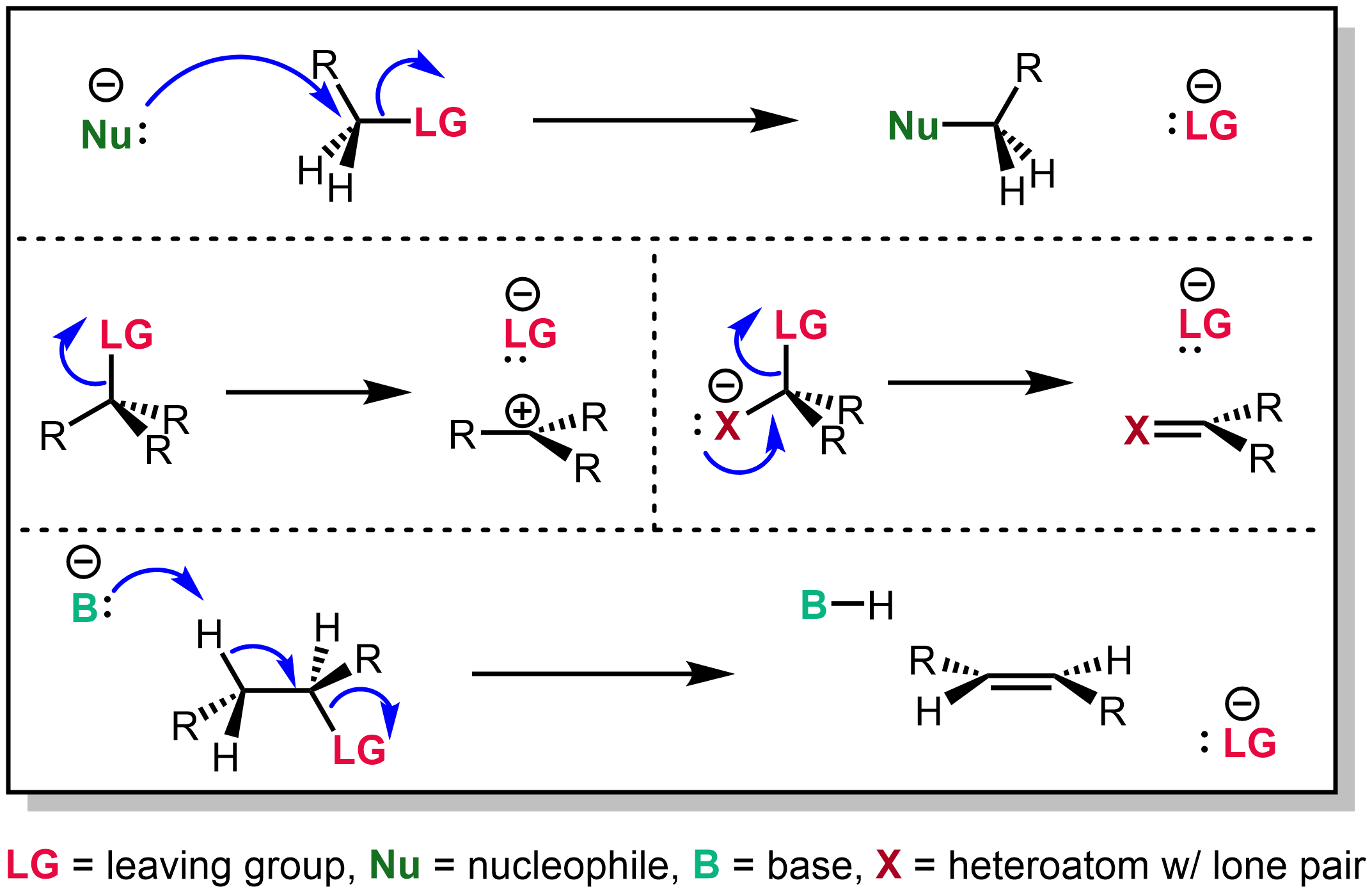
In chemistry, a nucleophile is a chemical species that forms bonds by donating an electron pair. All molecules and ions with a free pair of electrons or at least one pi bond can act as nucleophiles. Because nucleophiles donate electrons, they are Lewis bases. ''Nucleophilic'' describes the affinity of a nucleophile to bond with positively charged Atomic nucleus, atomic nuclei. Nucleophilicity, sometimes referred to as nucleophile strength, refers to a substance's nucleophilic character and is often used to compare the affinity of atoms. Neutral nucleophilic reactions with solvents such as Alcohol (chemistry), alcohols and water are named solvolysis. Nucleophiles may take part in nucleophilic substitution, whereby a nucleophile becomes attracted to a full or partial positive charge, and nucleophilic addition. Nucleophilicity is closely related to basicity. The difference between the two is, that basicity is a thermodynamic property (i.e. relates to an equilibrium state), but nucleop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SN1 Reaction
The unimolecular nucleophilic substitution (SN1) reaction is a substitution reaction in organic chemistry. The Hughes-Ingold symbol of the mechanism expresses two properties—"SN" stands for "nucleophilic substitution", and the "1" says that the rate-determining step is molecularity, unimolecular. Thus, the rate equation is often shown as having first-order dependence on the substrate and zero-order dependence on the nucleophile. This relationship holds for situations where the amount of nucleophile is much greater than that of the intermediate. Instead, the rate equation may be more accurately described using Steady state (chemistry), steady-state kinetics. The reaction involves a carbocation intermediate and is commonly seen in reactions of secondary or tertiary alkyl halides under strongly basic conditions or, under strongly acidic conditions, with Alcohol (chemistry), secondary or tertiary alcohols. With primary and secondary alkyl halides, the alternative SN2 reaction, SN2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leaving Group
In organic chemistry, a leaving group typically means a Chemical species, molecular fragment that departs with an electron, electron pair during a reaction step with heterolysis (chemistry), heterolytic bond cleavage. In this usage, a ''leaving group'' is a less formal but more commonly used synonym of the term ''nucleofuge''; although IUPAC gives the term a broader definition. A species' ability to serve as a leaving group can affect whether a reaction is viable, as well as what mechanism the reaction takes. Leaving group ability depends strongly on context, but often correlates with ability to stabilize additional electron density from bond heterolysis. Common anionic leaving groups are , and halides and sulfonate esters such as tosylate (). Water (), alcohols (), and amines () are common neutral leaving groups, although they often require activating catalysts. Some moieties, such as hydride (H−) serve as leaving groups only extremely rarely. Nomenclature IUPAC ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Schiff Base
In organic chemistry, a Schiff base (named after Hugo Schiff) is a compound with the general structure ( = alkyl or aryl, but not hydrogen). They can be considered a sub-class of imines, being either secondary ketimines or secondary aldimines depending on their structure. Anil refers to a common subset of Schiff bases: imines derived from anilines. The term can be synonymous with azomethine which refers specifically to secondary aldimines (i.e. where R' ≠ H). Synthesis Schiff bases can be synthesized from an aliphatic or aromatic amine and a carbonyl compound by nucleophilic addition forming a hemiaminal, followed by a dehydration to generate an imine. In a typical reaction, 4,4'-oxydianiline reacts with ''o''- vanillin: Schiff bases can also be synthesized via the Aza-Wittig reaction. Biochemistry Schiff bases have been investigated in relation to a wide range of contexts, including antimicrobial, antiviral and anticancer activity. They have also been con ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |