|
GlnALG Operon
The glnALG operon is an operon that regulates the nitrogen content of a cell. It codes for the structural gene glnA the two regulatory genes glnL and glnG. glnA encodes glutamine synthetase, an enzyme which catalyzes the conversion of glutamate and ammonia to glutamine, thereby controlling the nitrogen level in the cell. glnG encodes NRI which regulates the expression of the glnALG operon at three promoters, which are glnAp1, glnAp2 located upstream of glnA) and glnLp (intercistronic glnA-glnL region). glnL encodes NRII which regulates the activity of NRI. No significant homology is found in Eukaryotes. Structure The glnALG has three structural genes: * glnA: encodes glutamine synthetase, an enzyme which catalyzes the conversion of glutamate and ammonia to glutamine. * glnL: encodes NRII , which regulates the activity of NRI. * glnG: encodes NRI , which regulates the expression of glnALG operon at three promoters, which are glnAp1, glnAp2 and glnLp. Physiological significanc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Operon
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be ''co-transcribed'' to define an operon. Originally, operons were thought to exist solely in prokaryotes (which includes organelles like plastids that are derived from bacteria), but their discovery in eukaryotes was shown in the early 1990s, and are considered to be rare. In general, expression of prokaryotic operons leads to the generation of polycistronic mRNAs, while eukaryotic operons lead to monocistronic mRNAs. Operons are also found in viruses such as bacteriophages. For ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
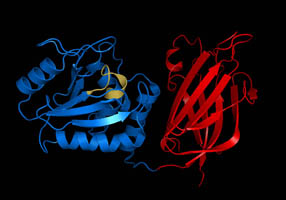
GlnALG Operon
The glnALG operon is an operon that regulates the nitrogen content of a cell. It codes for the structural gene glnA the two regulatory genes glnL and glnG. glnA encodes glutamine synthetase, an enzyme which catalyzes the conversion of glutamate and ammonia to glutamine, thereby controlling the nitrogen level in the cell. glnG encodes NRI which regulates the expression of the glnALG operon at three promoters, which are glnAp1, glnAp2 located upstream of glnA) and glnLp (intercistronic glnA-glnL region). glnL encodes NRII which regulates the activity of NRI. No significant homology is found in Eukaryotes. Structure The glnALG has three structural genes: * glnA: encodes glutamine synthetase, an enzyme which catalyzes the conversion of glutamate and ammonia to glutamine. * glnL: encodes NRII , which regulates the activity of NRI. * glnG: encodes NRI , which regulates the expression of glnALG operon at three promoters, which are glnAp1, glnAp2 and glnLp. Physiological significanc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dephosphorylation
In biochemistry, dephosphorylation is the removal of a phosphate () group from an organic compound by hydrolysis. It is a reversible post-translational modification. Dephosphorylation and its counterpart, phosphorylation, activate and deactivate enzymes by detaching or attaching phosphoric esters and anhydrides. A notable occurrence of dephosphorylation is the conversion of Adenosine triphosphate, ATP to Adenosine diphosphate, ADP and inorganic phosphate. Dephosphorylation employs a type of hydrolytic enzyme, or hydrolase, which cleaves ester bonds. The prominent hydrolase subclass used in dephosphorylation is phosphatase, which removes phosphate groups by hydrolysing phosphoric acid monoesters into a phosphate ion and a molecule with a free hydroxyl (–OH) group. The reversible phosphorylation-dephosphorylation reaction occurs in every physiological process, making proper function of protein phosphatases necessary for organism viability. Because protein dephosphorylation is a k ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Triphosphate
Adenosine triphosphate (ATP) is a nucleoside triphosphate that provides energy to drive and support many processes in living cell (biology), cells, such as muscle contraction, nerve impulse propagation, and chemical synthesis. Found in all known forms of life, it is often referred to as the "molecular unit of currency" for intracellular energy transfer. When consumed in a Metabolism, metabolic process, ATP converts either to adenosine diphosphate (ADP) or to adenosine monophosphate (AMP). Other processes regenerate ATP. It is also a Precursor (chemistry), precursor to DNA and RNA, and is used as a coenzyme. An average adult human processes around 50 kilograms (about 100 mole (unit), moles) daily. From the perspective of biochemistry, ATP is classified as a nucleoside triphosphate, which indicates that it consists of three components: a nitrogenous base (adenine), the sugar ribose, and the Polyphosphate, triphosphate. Structure ATP consists of three parts: a sugar, an amine base ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catabolite Activator Protein
In cell biology, catabolite activator protein (CAP), which is also known as cAMP receptor protein (CRP), is a trans-acting transcriptional activator in bacteria that effectively catalyzes the initiation of DNA transcription by interacting with RNA polymerase in a way that causes the DNA to bend. CAP's name reflects the protein's ability to affect transcription of genes involved in many catabolic pathways. For example, when the amount of glucose transported into a cell is low, a cascade of events results in the increase of cAMP levels in the cell's cytosol, and this increase in cAMP levels is sensed by CAP, which goes on to activate the transcription of many other catabolic genes. CAP exists as a homodimer in solution, and it is bound to by two cyclic AMP (cAMP) ligand molecules with negative cooperativity. By increasing CAP's affinity for DNA, cyclic AMP functions as an allosteric effector. With its cyclic-AMP ligand, CAP binds a DNA region upstream from the site at whic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Binding
Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other. It is formed when atoms or molecules bind together by sharing of electrons. It often, but not always, involves some chemical bonding. In some cases, the associations can be quite strong—for example, the protein streptavidin and the vitamin biotin have a dissociation constant (reflecting the ratio between bound and free biotin) on the order of 10−14—and so the reactions are effectively irreversible. The result of molecular binding is sometimes the formation of a molecular complex in which the attractive forces holding the components together are generally non-covalent, and thus are normally energetically weaker than covalent bonds. Molecular binding occurs in biological complexes (e.g., between pairs or sets of proteins, or between a protein and a small molecule ligand it binds) and also in abiologic chemic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Concentration
In chemistry, concentration is the abundance of a constituent divided by the total volume of a mixture. Several types of mathematical description can be distinguished: '' mass concentration'', '' molar concentration'', '' number concentration'', and '' volume concentration''. The concentration can refer to any kind of chemical mixture, but most frequently refers to solutes and solvents in solutions. The molar (amount) concentration has variants, such as normal concentration and osmotic concentration. Dilution is reduction of concentration, e.g. by adding solvent to a solution. The verb to concentrate means to increase concentration, the opposite of dilute. Etymology ''Concentration-'', ''concentratio'', action or an act of coming together at a single place, bringing to a common center, was used in post-classical Latin in 1550 or earlier, similar terms attested in Italian (1589), Spanish (1589), English (1606), French (1632). Qualitative description Often in informal, non- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyclic Adenosine Monophosphate
Cyclic adenosine monophosphate (cAMP, cyclic AMP, or 3',5'-cyclic adenosine monophosphate) is a second messenger, or cellular signal occurring within cells, that is important in many biological processes. cAMP is a derivative of adenosine triphosphate (ATP) and used for intracellular signal transduction in many different organisms, conveying the cAMP-dependent pathway. History Earl Sutherland of Vanderbilt University won a Nobel Prize in Physiology or Medicine in 1971 "for his discoveries concerning the mechanisms of the action of hormones", especially epinephrine, via second messengers (such as cyclic adenosine monophosphate, cyclic AMP). Synthesis The synthesis of cAMP is stimulated by trophic hormones that bind to receptors on the cell surface. cAMP levels reach maximal levels within minutes and decrease gradually over an hour in cultured cells. Cyclic AMP is synthesized from ATP by adenylate cyclase located on the inner side of the plasma membrane and anchored at v ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carbon Starvation
Carbon () is a chemical element; it has chemical symbol, symbol C and atomic number 6. It is nonmetallic and tetravalence, tetravalent—meaning that its atoms are able to form up to four covalent bonds due to its valence shell exhibiting 4 electrons. It belongs to group 14 of the periodic table. Carbon makes up about 0.025 percent of Earth's crust. Three Isotopes of carbon, isotopes occur naturally, carbon-12, C and carbon-13, C being stable, while carbon-14, C is a radionuclide, decaying with a half-life of 5,700 years. Carbon is one of the timeline of chemical element discoveries#Pre-modern and early modern discoveries, few elements known since antiquity. Carbon is the 15th abundance of elements in Earth's crust, most abundant element in the Earth's crust, and the abundance of the chemical elements, fourth most abundant element in the universe by mass after hydrogen, helium, and oxygen. Carbon's abundance, its unique diversity of organic compounds, and its unusual abi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |