|
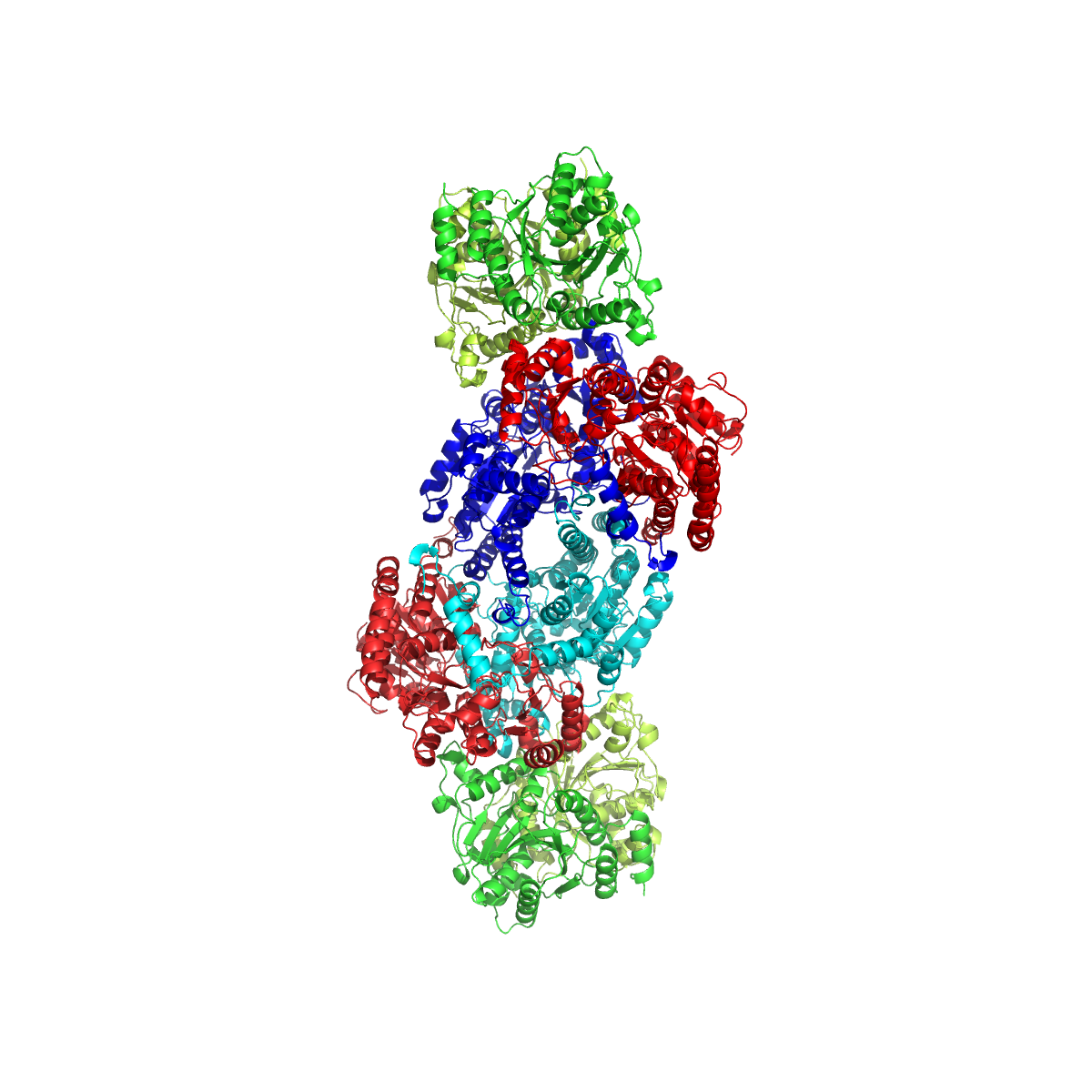
Dinitrogenase
Nitrogenases are enzymes () that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the Organic redox reaction, reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a key step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or Homologous chromosome, homologs. They are related to protochlorophyllide reductase. Classification and structure Although the equilibrium formation of ammonia from molecular hydrogen and nitrogen has an overall negative enthalpy of reaction ( \Delta H^ = -45.2 \ \mathrm \, \mathrm \; \mathrm ), the activation energy is very high ( E_\mathrm = 230-420 \ \mathrm \, \mathrm ). Nitrogenase a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nitrogenase
Nitrogenases are enzymes () that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a key step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules ( nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or homologs. They are related to protochlorophyllide reductase. Classification and structure Although the equilibrium formation of ammonia from molecular hydrogen and nitrogen has an overall negative enthalpy of reaction ( \Delta H^ = -45.2 \ \mathrm \, \mathrm \; \mathrm ), the activation energy is very high ( E_\mathrm = 230-420 \ \mathrm \, \mathrm ). Nitrogenase acts as a catalyst, reducing this energy barr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activation Energy
In chemistry and physics, activation energy is the minimum amount of energy that must be provided for compounds to result in a chemical reaction. The activation energy (''E''a) of a reaction is measured in joules per mole (J/mol), kilojoules per mole (kJ/mol) or kilocalories per mole (kcal/mol). Activation energy can be thought of as the magnitude of the potential barrier (sometimes called the energy barrier) separating minima of the potential energy surface pertaining to the initial and final thermodynamic state. For a chemical reaction to proceed at a reasonable rate, the temperature of the system should be high enough such that there exists an appreciable number of molecules with translational energy equal to or greater than the activation energy. The term "activation energy" was introduced in 1889 by the Swedish scientist Svante Arrhenius. Other uses Although less commonly used, activation energy also applies to nuclear reactions and various other physical phenomena. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FeMo Cofactor
FeMoco ( cofactor) is the primary cofactor of nitrogenase. Nitrogenase is the enzyme that catalyzes the conversion of atmospheric nitrogen molecules N2 into ammonia (NH3) through the process known as nitrogen fixation. Containing iron and molybdenum, the cofactor is called FeMoco. Its stoichiometry is Fe7MoS9C. Structure The FeMo cofactor is a cluster with composition Fe7MoS9C. Fe is the chemical symbol for the element iron (ferrum), and Mo is the symbol for molybdenum. This large cluster can be viewed as two subunits composed of one Fe4S3 (iron(III) sulfide) cluster and one MoFe3S3 cluster. The two clusters are linked by three sulfide ligands. The unique iron (Fe) is anchored to the protein by a cysteine. It is also bound to three sulfides, resulting in tetrahedral molecular geometry. The additional six Fe centers in the cluster are each bonded to three sulfides. These six internal Fe centers define a trigonal prismatic arrangement around a central carbide center. The molyb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Iron–sulfur Cluster
Iron–sulfur clusters (or iron–sulphur clusters in British spelling) are molecular ensembles of iron and sulfide. They are most often discussed in the context of the biological role for iron–sulfur proteins, which are pervasive. Many Fe–S clusters are known in the area of organometallic chemistry and as precursors to synthetic analogues of the biological clusters (see Figure). It is believed that the last universal common ancestor had many iron-sulfur clusters. Organometallic clusters Organometallic Fe–S clusters include the sulfido carbonyls with the formula Fe2S2(CO)6, H2Fe3S(CO)9, and Fe3S2(CO)9. Compounds are also known that incorporate cyclopentadienyl ligands, such as (C5H5)4Fe4S4. Inorganic materials center, Structure of potassium dithioferrate, which features infinite chains of Fe(III) centers. Biological Fe–S clusters Iron–sulfur clusters occur in many biological systems, often as components of electron transfer proteins. The ferredoxin protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Triphosphate
Adenosine triphosphate (ATP) is an organic compound that provides energy to drive many processes in living cells, such as muscle contraction, nerve impulse propagation, condensate dissolution, and chemical synthesis. Found in all known forms of life, ATP is often referred to as the "molecular unit of currency" of intracellular energy transfer. When consumed in metabolic processes, it converts either to adenosine diphosphate (ADP) or to adenosine monophosphate (AMP). Other processes regenerate ATP. The human body recycles its own body weight equivalent in ATP each day. It is also a precursor to DNA and RNA, and is used as a coenzyme. From the perspective of biochemistry, ATP is classified as a nucleoside triphosphate, which indicates that it consists of three components: a nitrogenous base ( adenine), the sugar ribose, and the triphosphate. Structure ATP consists of an adenine attached by the 9-nitrogen atom to the 1′ carbon atom of a sugar ( ribose), which in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
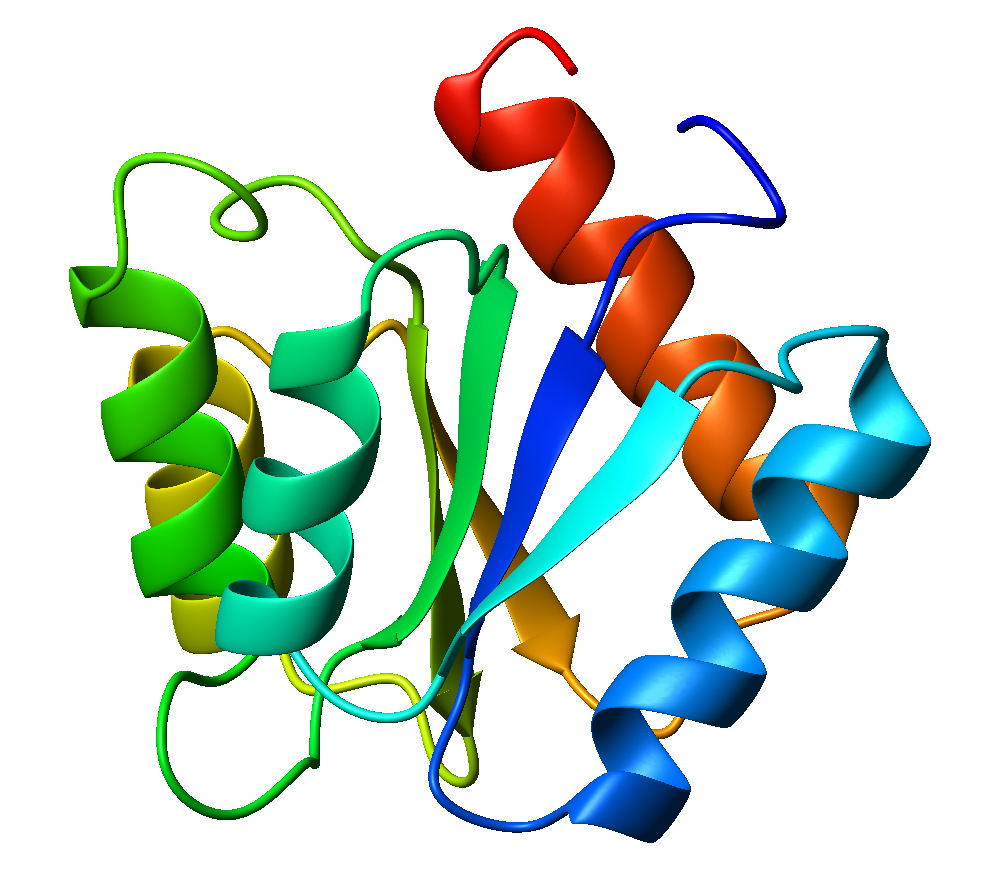
Flavodoxin
Flavodoxins (Fld) are small, soluble electron-transfer proteins. Flavodoxins contains flavin mononucleotide as prosthetic group. The structure of flavodoxin is characterized by a five-stranded parallel beta sheet, surrounded by five alpha helices. They have been isolated from prokaryotes, cyanobacteria, and some eukaryotic algae. Background Originally found in cyanobacteria and clostridia, flavodoxins were discovered over 50 years ago. These proteins evolved from an anaerobic environment, due to selective pressures. Ferredoxin, another redox protein, was the only protein able to be used in this manner. However, when oxygen became present in the environment, iron became limited. Ferredoxin is iron-dependant as well as oxidant-sensitive. Under these limited iron conditions, ferredoxin was no longer preferred. Flavodoxin on the other hand is the opposite of these traits, as it is oxidant-resistant and has iron-free isofunctional counterparts. Therefore, for some time flavodoxin wa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ferredoxin
Ferredoxins (from Latin ''ferrum'': iron + redox, often abbreviated "fd") are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium '' Clostridium pasteurianum''. Another redox protein, isolated from spinach chloroplasts, was termed "chloroplast ferredoxin". The chloroplast ferredoxin is involved in both cyclic and non-cyclic photophosphorylation reactions of photosynthesis. In non-cyclic photophosphorylation, ferredoxin is the last electron acceptor thus reducing the enzyme NADP+ reductase. It accepts electrons produced from sunlight- excited chlorophyll and transfers them to the enzyme ferredoxin: NADP+ oxidoreductase . Ferredoxins are small proteins containing iron and sulfur atoms organized as iron–sulfur clusters. These biological " capacitors" ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reducing Agent
In chemistry, a reducing agent (also known as a reductant, reducer, or electron donor) is a chemical species that "donates" an electron to an (called the , , , or ). Examples of substances that are commonly reducing agents include the Earth metals, formic acid, oxalic acid, and sulfite compounds. In their pre-reaction states, reducers have extra electrons (that is, they are by themselves reduced) and oxidizers lack electrons (that is, they are by themselves oxidized). This is commonly expressed in terms of their oxidation states. An agent's oxidation state describes its degree of loss of electrons, where the higher the oxidation state then the fewer electrons it has. So initially, prior to the reaction, a reducing agent is typically in one of its lower possible oxidation states; its oxidation state increases during the reaction while that of the oxidizer decreases. Thus in a redox reaction, the agent whose oxidation state increases, that "loses/ donates electrons", that "i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Reviews
''Chemical Reviews'' is peer-reviewed scientific journal published twice per month by the American Chemical Society. It publishes review articles on all aspects of chemistry. It was established in 1924 by William Albert Noyes ( University of Illinois). the editor-in-chief is Sharon Hammes-Schiffer. Abstracting and indexing The journal is abstracted and indexed in Chemical Abstracts Service, CAB International, EBSCOhost, ProQuest, PubMed, Scopus, and the Science Citation Index. According to the ''Journal Citation Reports'', the journal has a 2020 impact factor The impact factor (IF) or journal impact factor (JIF) of an academic journal is a scientometric index calculated by Clarivate that reflects the yearly mean number of citations of articles published in the last two years in a given journal, as ... of 60.622. See also * Accounts of Chemical Research References External links * American Chemical Society academic journals Review journals Monthly jo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FeMoco Cluster
FeMoco ( cofactor) is the primary cofactor of nitrogenase. Nitrogenase is the enzyme that catalyzes the conversion of atmospheric nitrogen molecules N2 into ammonia (NH3) through the process known as nitrogen fixation. Containing iron and molybdenum, the cofactor is called FeMoco. Its stoichiometry is Fe7MoS9C. Structure The FeMo cofactor is a cluster with composition Fe7MoS9C. Fe is the chemical symbol for the element iron (ferrum), and Mo is the symbol for molybdenum. This large cluster can be viewed as two subunits composed of one Fe4S3 (iron(III) sulfide) cluster and one MoFe3S3 cluster. The two clusters are linked by three sulfide ligands. The unique iron (Fe) is anchored to the protein by a cysteine. It is also bound to three sulfides, resulting in tetrahedral molecular geometry. The additional six Fe centers in the cluster are each bonded to three sulfides. These six internal Fe centers define a trigonal prismatic arrangement around a central carbide center. The molyb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reductase
A reductase is an enzyme that catalyzes a reduction reaction. Examples * 5α-Reductase * 5β-Reductase * Dihydrofolate reductase * HMG-CoA reductase * Methemoglobin reductase * Ribonucleotide reductase * Thioredoxin reductase * ''E. coli'' nitroreductase * Methylenetetrahydrofolate reductase See also * Oxidase * Oxidoreductase In biochemistry, an oxidoreductase is an enzyme that catalyzes the transfer of electrons from one molecule, the reductant, also called the electron donor, to another, the oxidant, also called the electron acceptor. This group of enzymes usually ... References Oxidoreductases {{Enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid resid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |