|
Tat (HIV)
In molecular biology, Tat is a protein that is encoded for by the ''tat'' gene in HIV-1. Tat is a regulatory protein that drastically enhances the efficiency of viral transcription. Tat stands for "Trans-Activator of Transcription". The protein consists of between 86 and 101 amino acids depending on the subtype. Tat vastly increases the level of transcription of the HIV dsDNA. Before Tat is present, a small number of RNA transcripts will be made, which allow the Tat protein to be produced. Tat then binds to cellular factors and mediates their phosphorylation, resulting in increased transcription of all HIV genes, providing a positive feedback cycle. This in turn allows HIV to have an explosive response once a threshold amount of Tat is produced, a useful tool for defeating the body's response. Tat also appears to play a more direct role in the HIV disease process. The protein is released by infected cells in culture, and is found in the blood of HIV-1 infected patients. It ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactions. Though cells and other microscopic structures had been observed in living organisms as early as the 18th century, a detailed understanding of the mechanisms and interactions governing their behavior did not emerge until the 20th century, when technologies used in physics and chemistry had advanced sufficiently to permit their application in the biological sciences. The term 'molecular biology' was first used in 1945 by the English physicist William Astbury, who described it as an approach focused on discerning the underpinnings of biological phenomena—i.e. uncovering the physical and chemical structures and properties of biological molecules, as well as their interactions with other molecules and how these interactions explain observ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
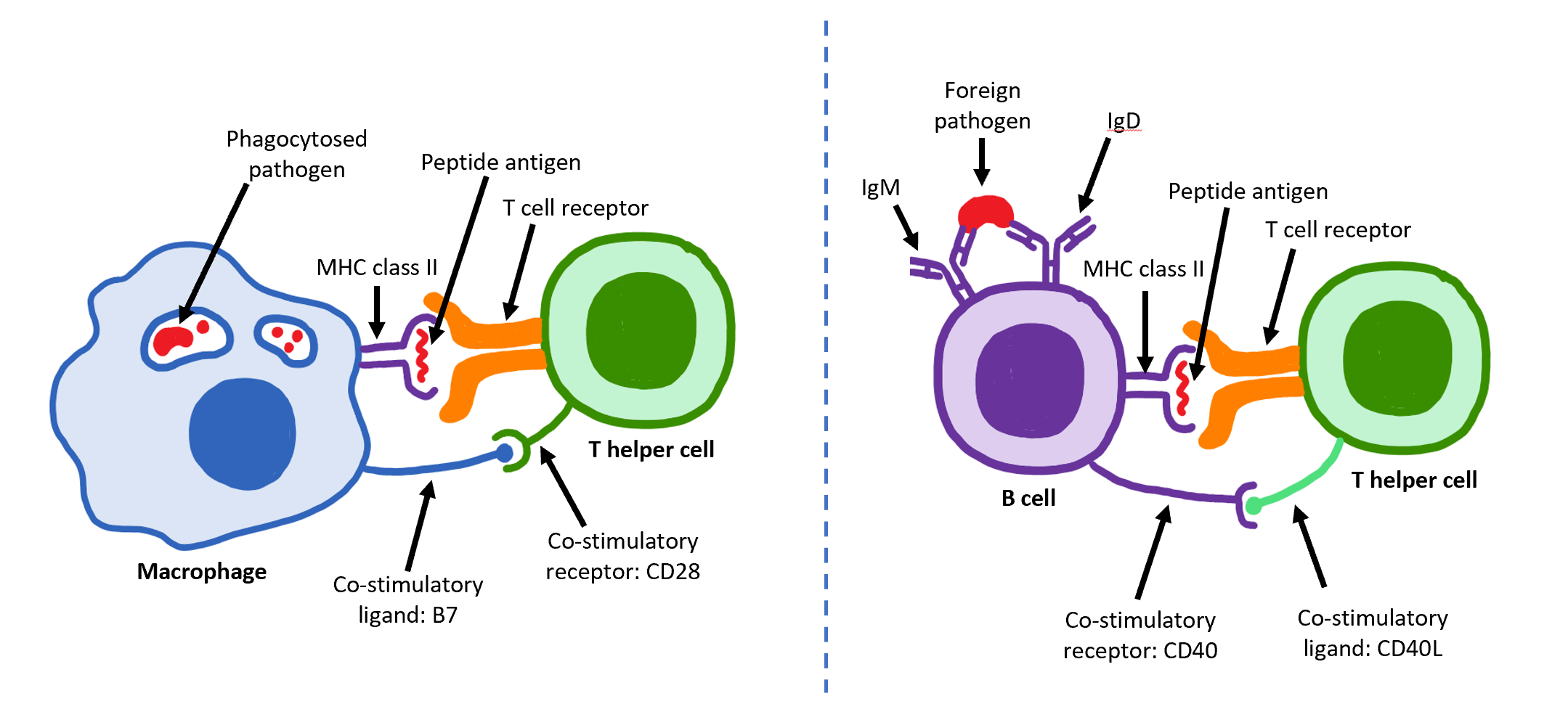
T Helper Cell
The T helper cells (Th cells), also known as CD4+ cells or CD4-positive cells, are a type of T cell that play an important role in the adaptive immune system. They aid the activity of other immune cells by releasing cytokines. They are considered essential in B cell Immunoglobulin class switching, antibody class switching, breaking Cross-presentation, cross-tolerance in dendritic cells, in the activation and growth of cytotoxic T cells, and in maximizing bactericidal activity of phagocytes such as macrophages and neutrophils. CD4+ cells are mature Th cells that express the surface protein CD4. Genetic variation in regulatory elements expressed by CD4+ cells determines susceptibility to a broad class of autoimmune diseases. Structure and function Th cells contain and release cytokines to aid other immune cells. Cytokines are small protein mediators that alter the behavior of target cells that express Receptor (biochemistry), receptors for those cytokines. These cells help polar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Overview For transcription to take place, the enzyme that synthesizes RNA, known as RNA polymerase, must attach to the DNA near a gene. Promoters contain specific DNA sequences such as response elements that provide a secure initial binding site for RNA polymerase and for proteins called transcription factors that recruit RNA polymerase. These transcription factor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase II
RNA polymerase II (RNAP II and Pol II) is a Protein complex, multiprotein complex that Transcription (biology), transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNA polymerase, RNAP enzymes found in the nucleus of eukaryote, eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoter (biology), promoters and begin transcription. Discovery Early studies suggested a minimum of two RNAPs: one which synthesized rRNA in the nucleolus, and one which synthesized other RNA in the nucleoplasm, part of the nucleus but outside the nucleolus. In 1969, biochemists Robert G. Roeder and William J. Rutter, William Rutter discovered there are total three distinct nuclear RNA polymerases, an additional RNAP that was responsible for transcription of some kind of RNA in the nucleoplasm. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyclin T1
Cyclin-T1 is a protein that in humans is encoded by the ''CCNT1'' gene. Function The protein encoded by this gene belongs to the highly conserved cyclin family, whose members are characterized by a dramatic periodicity in protein abundance through the cell cycle. Cyclins function as regulators of CDK kinases. Different cyclins exhibit distinct expression and degradation patterns that contribute to the temporal coordination of each mitotic event. This cyclin tightly associates with CDK9 kinase, and was found to be a major subunit of the transcription elongation factor p-TEFb. The kinase complex containing this cyclin and the elongation factor can interact with, and act as a cofactor of human immunodeficiency virus type 1 (HIV-1) Tat protein, and was shown to be both necessary and sufficient for full activation of viral transcription. This cyclin and its kinase partner were also found to be involved in the phosphorylation and regulation of the carboxy-terminal domain (CTD) of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CDK9
Cyclin-dependent kinase 9 or CDK9 is a cyclin-dependent kinase associated with P-TEFb. Function The protein encoded by this gene is a member of the cyclin-dependent kinase (CDK) family. CDK family members are highly similar to the gene products of S. cerevisiae cdc28, and S. pombe cdc2, and known as important cell cycle regulators. This kinase was found to be a component of the multiprotein complex TAK/P-TEFb, which is an elongation factor for RNA polymerase II-directed transcription and functions by phosphorylating the C-terminal domain of the largest subunit of RNA polymerase II. This protein forms a complex with and is regulated by its regulatory subunit cyclin T or cyclin K. HIV-1 Tat protein was found to interact with this protein and cyclin T, which suggested a possible involvement of this protein in AIDS. CDK9 is also known to associate with other proteins such as TRAF2, and be involved in differentiation of skeletal muscle. Inhibitors Based on molecular docking res ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P-TEFb
The positive transcription elongation factor, P-TEFb, is a multiprotein complex that plays an essential role in the regulation of transcription by RNA polymerase II (Pol II) in eukaryotes. Immediately following initiation Pol II becomes trapped in promoter proximal paused positions on the majority of human genes (Figure 1). P-TEFb is a cyclin dependent kinase that can phosphorylate the 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole, DRB sensitivity inducing factor (DSIF) and negative elongation factor (NELF), as well as the carboxyl terminal domain of the large subunit of Pol IIMarshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 1996; 271:27176-83. and this causes the transition into productive elongation leading to the synthesis of mRNAs. P-TEFb is regulated in part by a reversible association with the 7SK RNA, 7SK snRNP.Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that pl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Multiprotein Complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multidomain enzymes, in which multiple catalytic domains are found in a single polypeptide chain. Protein complexes are a form of quaternary structure. Proteins in a protein complex are linked by non-covalent protein–protein interactions. These complexes are a cornerstone of many (if not most) biological processes. The cell is seen to be composed of modular supramolecular complexes, each of which performs an independent, discrete biological function. Through proximity, the speed and selectivity of binding interactions between enzymatic complex and substrates can be vastly improved, leading to higher cellular efficiency. Many of the techniques used to enter cells and isolate proteins are inherently disruptive to such large complexes, complicating the task of determining the components of a complex. Examples of protein complexes include the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Directionality (molecular Biology)
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide Pentose, pentose-sugar-ring means that there will be a 5′ end (usually pronounced "five-prime end"), which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′ end (usually pronounced "three-prime end"), which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or Transcription (biology), transcription of the encoded information. Nucleic acids can only be synthesized in vivo in the 5′-to-3′ direction, as the polymerases that assemble various types of new strands generally rely on the energy produced by breaking nucleoside triphosphate bonds to attach new nucleo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Trans-activation Response Element
The HIV trans-activation response (TAR) element is an RNA element which is known to be required for the trans-activation of the viral promoter and for virus replication. The TAR hairpin is a dynamic structure that acts as a binding site for the Tat protein, and this interaction stimulates the activity of the long terminal repeat promoter. Further analysis has shown that TAR is a pre-microRNA that produces mature microRNAs from both strands of the TAR stem-loop. These miRNAs are thought to prevent infected cells from undergoing apoptosis by downregulating the genes ERCC1, IER3, CDK9, and Bim. Human polyomavirus 2 (JC virus Human polyomavirus 2, commonly referred to as the JC virus or John Cunningham virus, is a type of human polyomavirus. It was identified by electron microscopy in 1965 by ZuRhein and Chou, and by Silverman and Rubinstein. It was later isolated in ...) contains a TAR-homologous sequence in its late promoter that is responsive to HIV-1 derived Tat. Re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loops are nucleic acid Biomolecular structure, secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regions of the same nucleic acid strand, usually Complementarity (molecular biology), complementary in nucleotide sequence, base-pair to form a double helix that ends in a loop of unpaired nucleotides. Stem-loops are most commonly found in RNA, and are a key building block of many RNA biomolecular structure#Secondary structure, secondary structures. Stem-loops can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA-binding protein, RNA binding proteins, and serve as a Substrate (chemistry), substrate for Enzyme catalysis, enzymatic reactions. Formation and stability The formation of a stem-loop is dependent on the stability of the helix and loop regions. The first prerequisite is the p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Binding
Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other. It is formed when atoms or molecules bind together by sharing of electrons. It often, but not always, involves some chemical bonding. In some cases, the associations can be quite strong—for example, the protein streptavidin and the vitamin biotin have a dissociation constant (reflecting the ratio between bound and free biotin) on the order of 10−14—and so the reactions are effectively irreversible. The result of molecular binding is sometimes the formation of a molecular complex in which the attractive forces holding the components together are generally non-covalent, and thus are normally energetically weaker than covalent bonds. Molecular binding occurs in biological complexes (e.g., between pairs or sets of proteins, or between a protein and a small molecule ligand it binds) and also in abiologic chemic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |