|
TERF1
Telomeric repeat-binding factor 1 is a protein that in humans is encoded by the ''TERF1'' gene. Gene The human TERF1 gene is located in the chromosome 8 at 73,921,097-73,960,357 bp. Two transcripts of this gene are alternatively spliced products. The TERF1 gene is also known as TRF, PIN2 (Proteinase Inhibitor 2), TRF1, t-TRF1 and h-TRF1-AS. Protein The protein structure contains a C-terminal Myb motif, a dimerization domain (TERF homology) near its N-terminus and an acidic N-terminus. Subcellular distribution The cellular distribution of this DNA binding protein features the nucleoplasm, chromosomes, a telomeric region, a nuclear telomere cap complex, the cytoplasm, the spindle, the nucleus and a nucleolus and a nuclear chromosome. Function TERF 1 gene encodes a telomere specific protein which is a component of the telomere's shelterin nucleoprotein complex. This protein is present at telomeres throughout the cell cycle and functions as an inhibitor of telomer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TINF2
TERF1-interacting nuclear factor 2 is a protein that in humans is encoded by the ''TINF2'' gene. TINF2 is a component of the shelterin protein complex found at the end of telomeres. Interactions TINF2 has been shown to interact with ACD, POT1 Protection of telomeres protein 1 is a protein that in humans is encoded by the ''POT1'' gene. Function This gene is a member of the telombin family and encodes a nuclear protein involved in telomere maintenance. Specifically, this protein fu ... and TERF1. References Further reading * * * * * * * * * * * * * * * * External links GeneReviews/NCBI/NIH/UW entry on Dyskeratosis CongenitaPDBe-KBprovides an overview of all the structure information available in the PDB for Human TERF1-interacting nuclear factor 2 (TINF2) Telomere-related proteins {{gene-14-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PINX1
''PIN2/TERF1''-interacting telomerase inhibitor 1, also known as ''PINX1'', is a human gene. ''PINX1'' is also known as ''PIN2'' interacting protein 1. ''PINX1'' is a telomerase inhibitor and a possible tumor suppressor. Interactions PINX1 has been shown to interact with MCRS1, TERF1 and telomerase reverse transcriptase. Structure There are two known variants of PINX1. The second variant “lacks an exon in the 3’ coding region which results in a frameshift compared to variant 1. The encoded isoform is shorter and has a distinct C-terminus compared to isoform 1.” There are three ''PINX1'' cDNA clones. The longest one encodes a 328 amino acid 45kDa protein which contains an N-terminal Gly-rich patch and a C-terminal TID domain (telomerase inhibitory domain). The TRF1 binding domain is in the C-terminal 75 amino acids of ''PINX1''. Mouse PINX1 is 74% identical to human PINX1. In other eukaryotes, including yeast, there is an overall 50% similarity to human PINX1. Function ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TNKS
Tankyrase, also known as tankyrase 1, is an enzyme that in humans is encoded by the ''TNKS'' gene. It inhibits the binding of TERF1 to telomeric DNA. Tankyrase attracts substantial interest in cancer research through its interaction with AXIN1 and AXIN2, which are negative regulators of pro-oncogenic β-catenin signaling. Importantly, activity in the β-catenin destruction complex can be increased by tankyrase inhibitors and thus such inhibitors are a potential therapeutic option to reduce the growth of β-catenin-dependent cancers. Description Tankyrase-1 is a poly-ADP-ribosyltransferase involved in various processes such as Wnt signaling pathway, telomere length and vesicle trafficking. Acts as an activator of the Wnt signaling pathway by mediating poly-ADP-ribosylation (PARylation) of AXIN1 and AXIN2, 2 key components of the beta-catenin destruction complex: poly-ADP-ribosylated target proteins are recognized by RNF146, which mediates their ubiquitination and subsequent degr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SALL1
Sal-like 1 (Drosophila), also known as SALL1, is a protein which in humans is encoded by the ''SALL1'' gene. As the full name suggests, it is one of the human versions of the ''spalt'' (''sal'') gene known in ''Drosophila''. Function The protein encoded by this gene is a zinc finger transcriptional repressor and may be part of the NuRD histone deacetylase (HDAC) complex. Clinical significance Defects in this gene are a cause of Townes–Brocks syndrome (TBS) as well as branchio-oto-renal syndrome (BOR). Two transcript variants encoding different isoforms have been found for this gene. Interactions SALL1 has been shown to interact with TERF1 and UBE2I SUMO-conjugating enzyme UBC9 is an enzyme that in humans is encoded by the ''UBE2I'' gene. It is also sometimes referred to as "ubiquitin conjugating enzyme E2I" or "ubiquitin carrier protein 9", even though these names do not accurately describe .... References External links GeneReviews/NCBI/NIH/UW entry on T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAPRE1
Microtubule-associated protein RP/EB family member 1 is a protein that in humans is encoded by the ''MAPRE1'' gene. Function The protein encoded by this gene was first identified by its binding to the APC (Adenomatous polyposis coli) protein which is often mutated in familial and sporadic forms of colorectal cancer. Immunofluorescence has demonstrated that EB1 localizes to the centrosome, mitotic spindle, and distal tips of cytoplasmic microtubules. Throughout the cell cycle, EB1 proteins situate on the microtubule plus ends, which is why EB1 is categorized as a microtubule plus end tracking protein(+TIP protein). The protein also associates with components of the dynactin complex and the intermediate chain of cytoplasmic dynein. Because of these associations, it is thought that this protein is involved in the regulation of microtubule structures and chromosome stability. This gene is a member of the RP/EB family. Interactions MAPRE1 has been shown to interact with TERF ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Abl Gene
Tyrosine-protein kinase ABL1 also known as ABL1 is a protein that, in humans, is encoded by the ''ABL1'' gene (previous symbol ''ABL'') located on chromosome 9. c-Abl is sometimes used to refer to the version of the gene found within the mammalian genome, while v-Abl refers to the viral gene, which was initially isolated from the Abelson murine leukemia virus. Function The ''ABL1'' proto-oncogene encodes a cytoplasmic and nuclear protein tyrosine kinase that has been implicated in processes of cell differentiation, cell division, cell adhesion, and stress response such as DNA repair. Activity of ABL1 protein is negatively regulated by its SH3 domain, and deletion of the SH3 domain turns ABL1 into an oncogene. The t(9;22) translocation results in the head-to-tail fusion of the '' BCR'' and ''ABL1'' genes, leading to a fusion gene present in many cases of chronic myelogenous leukemia. The DNA-binding activity of the ubiquitously expressed ABL1 tyrosine kinase is regul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NME1
Nucleoside diphosphate kinase A is an enzyme that in humans is encoded by the ''NME1'' gene. It is thought to be a metastasis suppressor. Function This gene (NME1) was identified because of its reduced mRNA transcript levels in highly metastatic cells. Nucleoside diphosphate kinase (NDK) exists as a hexamer composed of 'A' (encoded by this gene) and 'B' (encoded by NME2) isoforms. Mutations in this gene have been identified in aggressive neuroblastomas. Two transcript variants encoding different isoforms have been found for this gene. Co-transcription of this gene and the neighboring downstream gene (NME2) generates naturally occurring transcripts (NME1-NME2), which encodes a fusion protein consisting of sequence sharing identity with each individual gene product. A bioinformatics study published in 2023 suggested that the ''NME1'' gene might have a prognostic role in neuroblastoma. Interactions NME1 has been shown to interact with: * Aurora A kinase, * CD29 * NME3 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TNKS2
Tankyrase-2 is an enzyme that in humans is encoded by the ''TNKS2'' gene In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei .... Interactions TNKS2 has been shown to interact with GRB14, TERF1 and Cystinyl aminopeptidase. References Further reading * * * * * * * * * * * * {{Gene-10-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
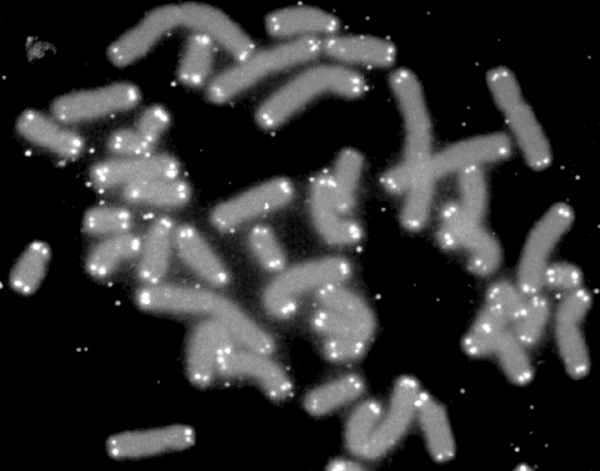
Telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes (see #Sequences, Sequences). Telomeres are a widespread genetic feature most commonly found in eukaryotes. In most, if not all species possessing them, they protect the terminal regions of DNA, chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the very ends of the DNA strand for a double-strand break. Discovery The existence of a special structure at the ends of chromosomes was independently proposed in 1938 by Hermann Joseph Muller, studying the fruit fly ''Drosophila melanogaster'', and in 1939 by Barbara McClintock, working with maize. Muller observed that the ends of irradiated fruit fly chromosomes did not present alterations such as deletions or inversions. He hypothesized the presence of a protective cap, which he coined "telomeres", from the Greek ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
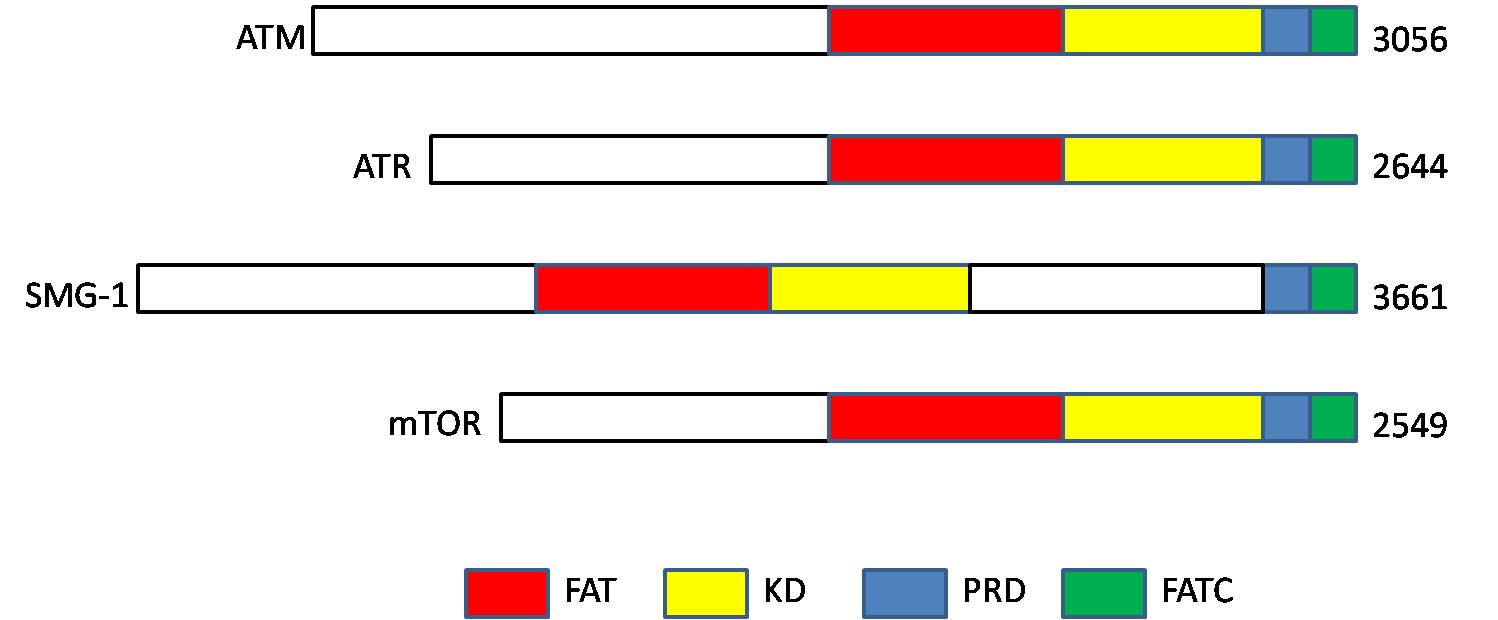
Ataxia Telangiectasia Mutated
ATM serine/threonine kinase or Ataxia-telangiectasia mutated, symbol ATM, is a serine/threonine protein kinase that is recruited and activated by DNA repair#Double-strand breaks, DNA double-strand breaks (Canonical pathway, canonical pathway), oxidative stress, topoisomerase cleavage complexes, splicing intermediates, R-loops and in some cases by DNA repair, single-strand DNA breaks. It phosphorylates several key proteins that initiate activation of the DNA damage cell cycle checkpoint, checkpoint, leading to cell cycle arrest, DNA repair or apoptosis. Several of these targets, including p53, CHK2, BRCA1, NBS1 and H2AX are tumor suppressors. In 1995, the gene was discovered by Yosef Shiloh who named its product ATM since he found that its mutations are responsible for the disorder ataxia–telangiectasia#Cause, ataxia–telangiectasia. In 1998, the Shiloh and Michael B. Kastan, Kastan laboratories independently showed that ATM is a protein kinase whose activity is enhanced by DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shelterin
Shelterin (also called telosome) is a protein complex known to protect telomeres in many eukaryotes from DNA repair mechanisms, as well as to regulate telomerase activity. In mammals and other vertebrates, telomeric DNA consists of repeating double-stranded 5'-TTAGGG-3' (G-strand) sequences (2-15 kilobases in humans) along with the 3'-AATCCC-5' (C-strand) complement, ending with a 50-400 nucleotide 3' (G-strand) overhang. Much of the final double-stranded portion of the telomere forms a T-loop (Telomere-loop) that is invaded by the 3' (G-strand) overhang to form a small D-loop (Displacement-loop). The absence of shelterin causes telomere uncapping and thereby activates damage-signaling pathways that may lead to non-homologous end joining (NHEJ), homology directed repair (HDR), end-to-end fusions, genomic instability, senescence, or apoptosis. Subunits Shelterin has six subunits: TRF1, TRF2, POT1, RAP1, TIN2, and TPP1. They can operate in smaller subsets to r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ABL (gene)
Tyrosine-protein kinase ABL1 also known as ABL1 is a protein that, in humans, is encoded by the ''ABL1'' gene (previous symbol ''ABL'') located on chromosome 9. c-Abl is sometimes used to refer to the version of the gene found within the mammalian genome, while v-Abl refers to the viral gene, which was initially isolated from the Abelson murine leukemia virus. Function The ''ABL1'' Oncogene#Proto-oncogene, proto-oncogene encodes a cytoplasmic and nuclear protein tyrosine kinase that has been implicated in processes of cell differentiation, cell division, cell adhesion, and stress response such as DNA repair. Activity of ABL1 protein is negatively regulated by its SH3 domain, and deletion of the SH3 domain turns ABL1 into an oncogene. The Philadelphia chromosome, t(9;22) translocation results in the head-to-tail fusion gene, fusion of the ''BCR gene, BCR'' and ''ABL1'' genes, leading to a fusion gene present in many cases of chronic myelogenous leukemia. The DNA-binding activit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |