|
TEAD2
TEAD2 (ETF, ETEF-1, TEF-4), together with TEAD1, defines a novel family of transcription factors, the TEAD family, highly conserved through evolution. TEAD proteins were notably found in ''Drosophila'' (Scalloped), ''C. elegans'' (egl -44), '' S. cerevisiae'' and '' A. nidulans''. TEAD2 has been less studied than TEAD1 but a few studies revealed its role during development. Function TEAD2 is a member of the mammalian TEAD transcription factor family (initially named the transcriptional enhancer factor (TEF) family), which contain the TEA/ATTS DNA-binding domain. Members of the family in mammals are TEAD1, TEAD2, TEAD3, TEAD4. Tissue distribution TEAD2 is selectively expressed in a subset of embryonic tissues including the cerebellum, testis, and distal portions of the forelimb and hindlimb buds, as well as the tail bud, but it is essentially absent from adult tissues. TEAD2 has also been shown to be expressed very early during development, i.e. from the 2-cell stage. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WWTR1
WW domain-containing transcription regulator protein 1 (WWTR1), also known as Transcriptional coactivator with PDZ-binding motif (TAZ), is a protein that in humans is encoded by the ''WWTR1'' gene. WWTR1 acts as a transcriptional coregulator and has no effect on transcription alone. When in complex with transcription factor binding partners, WWTR1 helps promote gene expression in pathways associated with development, cell growth and survival, and inhibiting apoptosis. Aberrant WWTR1 function has been implicated for its role in driving cancers. WWTR1 is often referred to as TAZ due to its initial characterization with the name TAZ. However, WWTR1 (TAZ) is not to be confused with the protein tafazzin, which originally held the official gene symbol TAZ, and is now TAFAZZIN. Structure WWTR1 contains a proline rich region, TEAD binding motif, WW domain, coiled coil region, and a transactivation domain (TAD) containing the PDZ domain-binding motif. WWTR1 (TAZ) lacks a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TEAD3
Transcriptional enhancer factor TEF-5 is a protein that in humans is encoded by the ''TEAD3'' gene. Function This gene product is a member of the transcriptional enhancer factor (TEF) family of transcription factors, which contain the TEA/ATTS DNA-binding domain. Members of the family in mammals are TEAD1, TEAD2, TEAD3, TEAD4. Transcriptional coregulators, such as WWTR1 (TAZ) bind to these transcription factors. TEAD3 is predominantly expressed in the placenta and is involved in the transactivation In the context of gene regulation: transactivation is the increased rate of gene expression triggered either by biological processes or by artificial means, through the expression of an intermediate transactivator protein. In the context of recep ... of the chorionic somatomammotropin-B gene enhancer. It is expressed in nervous system and muscle in fish embryos. Translation of this protein is initiated at a non-AUG (AUA) start codon. References Further reading * * * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TEAD4
Transcriptional enhancer factor TEF-3 is a protein that in humans is encoded by the ''TEAD4'' gene In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b .... Function This gene product is a member of the transcriptional enhancer factor (TEF) family of transcription factors, which contain the TEA/ATTS DNA-binding domain. Members of the family in mammals are TEAD1, TEAD2, TEAD3, TEAD4. TEAD4 is preferentially expressed in the skeletal muscle, and binds to the M-CAT regulatory element found in promoters of muscle-specific genes to direct their gene expression. Alternatively spliced transcripts encoding distinct isoforms, some of which are translated through the use of a non-AUG (UUG) initiation codon, have been described for this gene. Gene ablation experiments in mice (i.e. knockout mi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
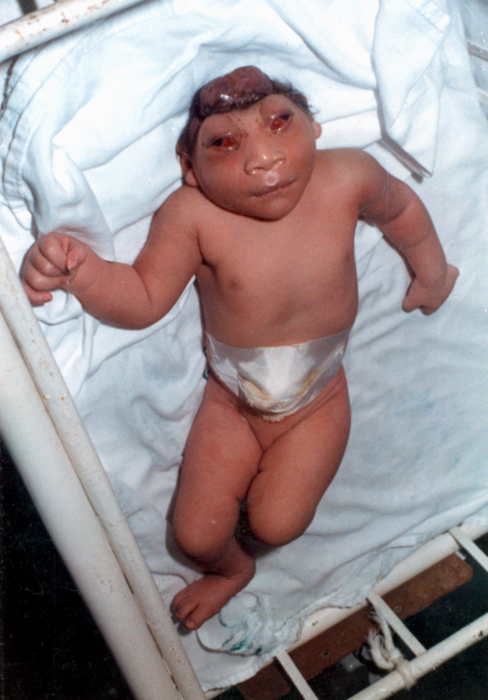
Anencephaly
Anencephaly is the absence of a major portion of the brain, skull, and scalp that occurs during embryonic development. It is a cephalic disorder that results from a neural tube defect that occurs when the rostral (head) end of the neural tube fails to close, usually between the 23rd and 26th day following conception. Strictly speaking, the Greek term translates as "without a brain" (or totally lacking the inside part of the head), but it is accepted that children born with this disorder usually only lack a telencephalon, the largest part of the brain consisting mainly of the cerebral hemispheres, including the neocortex, which is responsible for cognition. The remaining structure is usually covered only by a thin layer of membrane—skin, bone, meninges, etc., are all lacking. With very few exceptions, infants with this disorder do not survive longer than a few hours or days after birth. Signs and symptoms The National Institute of Neurological Disorders and Stroke (NINDS) desc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
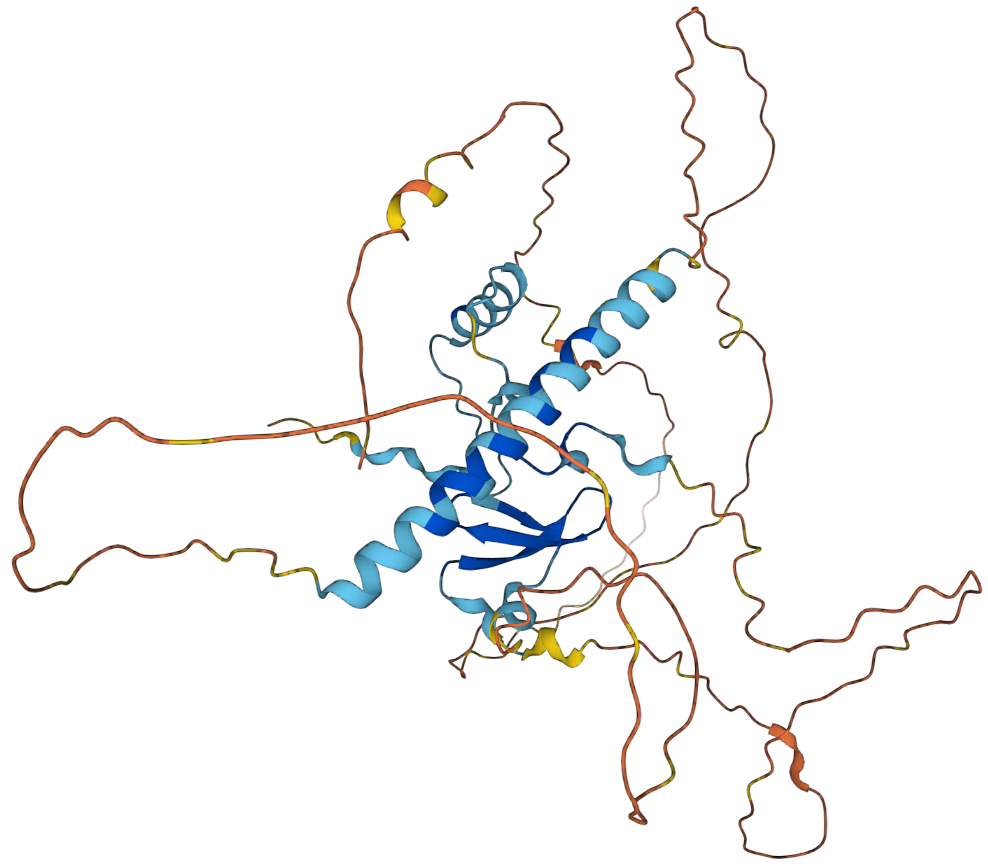
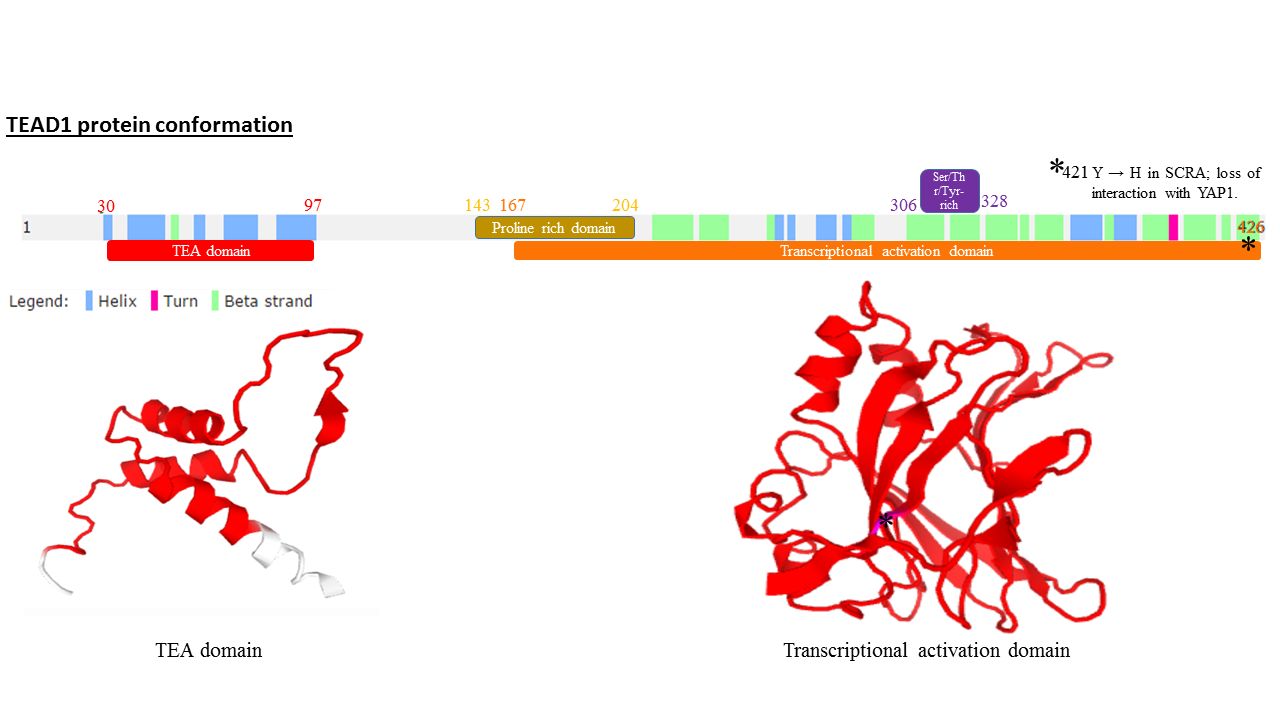
TEAD1
Transcriptional enhancer factor TEF-1 also known as TEA domain family member 1 (TEAD1) and transcription factor 13 (TCF-13) is a protein that in humans is encoded by the ''TEAD1'' gene. TEAD1 was the first member of the TEAD family of transcription factors to be identified. Structure All members of the TEAD family share a highly conserved DNA binding domain called the TEA domain. This DNA binding domain has a consensus DNA sequence 5’-CATTCCA/T-3’ that is called the MCAT element. The three dimensional structure of the TEA domain has been identified Its conformation is close to that of the homeodomain and contains 3 α helixes (H1, H2 and H3). It is the H3 helix that enables TEAD proteins to bind DNA. Another conserved domain of TEAD1 is located at the C terminus of the protein. It allows the binding of cofactors and has been called the YAP1 binding domain, because it is its ability to bind this well-known TEAD proteins co-factor that led to its identification. Indeed, T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization ( body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conserved Sequence
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domain (biology), domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA#History, DNA in heredity, and observations by Frederick Sanger of variation betwee ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila
''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species to linger around overripe or rotting fruit. They should not be confused with the Tephritidae, a related family, which are also called fruit flies (sometimes referred to as "true fruit flies"); tephritids feed primarily on unripe or ripe fruit, with many species being regarded as destructive agricultural pests, especially the Mediterranean fruit fly. One species of ''Drosophila'' in particular, '' D. melanogaster'', has been heavily used in research in genetics and is a common model organism in developmental biology. The terms "fruit fly" and "''Drosophila''" are often used synonymously with ''D. melanogaster'' in modern biological literature. The entire genus, however, contains more than 1,500 species and is very diverse in appea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caenorhabditis Elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek ''caeno-'' (recent), ''rhabditis'' (rod-like) and Latin ''elegans'' (elegant). In 1900, Maupas initially named it ''Rhabditides elegans.'' Osche placed it in the subgenus ''Caenorhabditis'' in 1952, and in 1955, Dougherty raised ''Caenorhabditis'' to the status of genus. ''C. elegans'' is an unsegmented pseudocoelomate and lacks respiratory or circulatory systems. Most of these nematodes are hermaphrodites and a few are males. Males have specialised tails for mating that include spicules. In 1963, Sydney Brenner proposed research into ''C. elegans,'' primarily in the area of neuronal development. In 1974, he began research into the molecular and developmental biology of ''C. elegans'', which has since been extensively used as a model organism. It was the first multice ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aspergillus Nidulans
''Aspergillus nidulans'' (also called ''Emericella nidulans'' when referring to its sexual form, or teleomorph) is one of many species of filamentous fungi in the phylum Ascomycota. It has been an important research organism for studying eukaryotic cell biology for over 50 years, being used to study a wide range of subjects including recombination, DNA repair, mutation, cell cycle control, tubulin, chromatin, nucleokinesis, pathogenesis, metabolism, and experimental evolution. It is one of the few species in its genus able to form sexual spores through meiosis, allowing crossing of strains in the laboratory. ''A. nidulans'' is a homothallic fungus, meaning it is able to self-fertilize and form fruiting bodies in the absence of a mating partner. It has septate hyphae with a woolly colony texture and white mycelia. The green colour of wild-type colonies is due to pigmentation of the spores, while mutations in the pigmentation pathway can produce other spore colours. Genome Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |