|
Stirrup Protein Domain
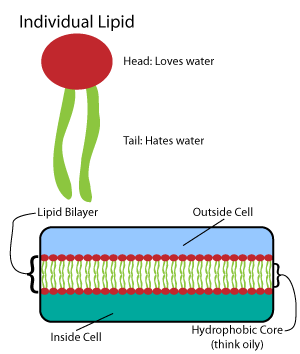
In molecular biology, the Stirrup is a protein domain found only in the domain archaea, in prokaryotic protein ribonucleotide reductases. It obtains its name due to its resemblance to an old fashioned Japanese stirrup. Stirrip has a molecular mass of 9 kDa and is folded into an alpha/beta structure. It allows for binding of the reductase to DNA via electrostatic interactions, since it has a predominance of positive charges distributed on its surface. Function This protein domain provides the precursors necessary for DNA synthesis. It catalyses the biosynthesis of DNA from RNA. Structure This structure contains a three-stranded beta-sheet to the solvent, which lies against alpha-helices An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix). The alpha helix is the most common structural arrangement in the secondary structure of proteins. It is also the most extreme type of l .... References {{InterPro content, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactions. Though cells and other microscopic structures had been observed in living organisms as early as the 18th century, a detailed understanding of the mechanisms and interactions governing their behavior did not emerge until the 20th century, when technologies used in physics and chemistry had advanced sufficiently to permit their application in the biological sciences. The term 'molecular biology' was first used in 1945 by the English physicist William Astbury, who described it as an approach focused on discerning the underpinnings of biological phenomena—i.e. uncovering the physical and chemical structures and properties of biological molecules, as well as their interactions with other molecules and how these interactions explain observ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding (molecular)
Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other. It is formed when atoms or molecules bind together by sharing of electrons. It often, but not always, involves some chemical bonding. In some cases, the associations can be quite strong—for example, the protein streptavidin and the vitamin biotin have a dissociation constant (reflecting the ratio between bound and free biotin) on the order of 10−14—and so the reactions are effectively irreversible. The result of molecular binding is sometimes the formation of a molecular complex in which the attractive forces holding the components together are generally non-covalent, and thus are normally energetically weaker than covalent bonds. Molecular binding occurs in biological complexes (e.g., between pairs or sets of proteins, or between a protein and a small molecule ligand it binds) and also in abiologic chemic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domains
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer after ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix). The alpha helix is the most common structural arrangement in the Protein secondary structure, secondary structure of proteins. It is also the most extreme type of local structure, and it is the local structure that is most easily predicted from a sequence of amino acids. The alpha helix has a right-handed helix conformation in which every backbone amino, N−H group hydrogen bonds to the backbone carbonyl, C=O group of the amino acid that is four residue (biochemistry), residues earlier in the protein sequence. Other names The alpha helix is also commonly called a: * Pauling–Corey–Branson α-helix (from the names of three scientists who described its structure) * 3.613-helix because there are 3.6 amino acids in one ring, with 13 atoms being involved in the ring formed by the hydrogen bond (starting with amidic hydrogen and ending with carbonyl oxygen) Discovery ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta-sheet
The beta sheet (β-sheet, also β-pleated sheet) is a common structural motif, motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone chain, backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of peptide, polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformational isomerism, conformation. The supramolecular association of β-sheets has been implicated in the formation of the Amyloid fibril, fibrils and Amyloid plaques, protein aggregates observed in amyloidosis, Alzheimer's disease and other Proteinopathy, proteinopathies. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biosynthesis
Biosynthesis, i.e., chemical synthesis occurring in biological contexts, is a term most often referring to multi-step, enzyme-Catalysis, catalyzed processes where chemical substances absorbed as nutrients (or previously converted through biosynthesis) serve as enzyme substrate (chemistry), substrates, with conversion by the living organism either into simpler or more complex Product (chemistry), products. Examples of biosynthetic pathways include those for the production of amino acids, lipid membrane components, and nucleotides, but also for the production of all classes of biological macromolecules, and of acetyl-coenzyme A, adenosine triphosphate, nicotinamide adenine dinucleotide and other key intermediate and transactional molecules needed for metabolism. Thus, in biosynthesis, any of an array of Chemical compound, compounds, from simple to complex, are converted into other compounds, and so it includes both the catabolism and anabolism (building up and breaking down) of comple ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Precursor (chemistry)
In chemistry, a precursor is a compound that participates in a chemical reaction that produces another compound. In biochemistry Biochemistry, or biological chemistry, is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology, a ..., the term "precursor" often refers more specifically to a chemical compound preceding another in a metabolic pathway, such as a protein precursor. Illicit drug precursors In 1988, the United Nations Convention Against Illicit Traffic in Narcotic Drugs and Psychotropic Substances introduced detailed provisions and requirements relating the control of precursors used to produce drugs of abuse. In Europe the Regulation (EC) No. 273/2004 of the European Parliament and of the Council on drug precursors was adopted on 11 February 2004. ( European law on drug precursors) Illicit explosives precursors On Ja ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Positive Predictive Value
The positive and negative predictive values (PPV and NPV respectively) are the proportions of positive and negative results in statistics and diagnostic tests that are true positive and true negative results, respectively. The PPV and NPV describe the performance of a diagnostic test or other statistical measure. A high result can be interpreted as indicating the accuracy of such a statistic. The PPV and NPV are not intrinsic to the test (as true positive rate and true negative rate are); they depend also on the prevalence. Both PPV and NPV can be derived using Bayes' theorem. Although sometimes used synonymously, a ''positive predictive value'' generally refers to what is established by control groups, while a post-test probability refers to a probability for an individual. Still, if the individual's pre-test probability of the target condition is the same as the prevalence in the control group used to establish the positive predictive value, the two are numerically equal. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electrostatic
Electrostatics is a branch of physics that studies slow-moving or stationary electric charges. Since classical times, it has been known that some materials, such as amber, attract lightweight particles after rubbing. The Greek word (), meaning 'amber', was thus the root of the word ''electricity''. Electrostatic phenomena arise from the forces that electric charges exert on each other. Such forces are described by Coulomb's law. There are many examples of electrostatic phenomena, from those as simple as the attraction of plastic wrap to one's hand after it is removed from a package, to the apparently spontaneous explosion of grain silos, the damage of electronic components during manufacturing, and photocopier and laser printer operation. The electrostatic model accurately predicts electrical phenomena in "classical" cases where the velocities are low and the system is macroscopic so no quantum effects are involved. It also plays a role in quantum mechanics, where addition ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reductase
In biochemistry, an oxidoreductase is an enzyme that catalyzes the transfer of electrons from one molecule, the reductant, also called the electron donor, to another, the oxidant, also called the electron acceptor. This group of enzymes usually utilizes NADP+ or NADH, NAD+ as Cofactor (biochemistry), cofactors. Transmembrane oxidoreductases create electron transport chains in bacteria, Photosystem, chloroplasts and mitochondria, including respiratory complexes Respiratory complex I, I, complex II, II and complex III, III. Some others can associate with biological membranes as peripheral membrane proteins or be anchored to the membranes through a single transmembrane helix. in Membranome database Reactions For example, an enzyme that ...[...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or Disulfide bond, disulfide bridges. Domains often form functional units, such as the calcium-binding EF-hand, EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimera (protein), chimeric ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |