|
Stemloc
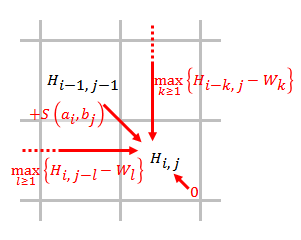
In bioinformatics, Stemloc is an open source software for multiple RNA sequence alignment and RNA structure prediction based on probabilistic models of RNA structure known as Pair stochastic context-free grammars (also probabilistic context-free grammars). Stemloc attempts to simultaneously predict and align the structure of RNA sequences with an improved time and space cost compared to previous methods with the same motive. The resulting software implements constrained versions of thSankoff algorithmby introducing both fold and alignment constraints, which reduces processor and memory usage and allows for larger RNA sequences to be analyzed on commodity hardware. Stemloc was written in 2004 by Ian Holmes. Stemloc can be downloaded as part of thDART software package It accepts input files in either FASTA or Stockholm format. Terminology * Fold: RNA folding is the process by which an RNA molecule acquires Protein secondary structure">secondary structure through intra-mo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Structure Prediction
Nucleic acid structure prediction is a computational method to determine ''secondary'' and ''tertiary'' nucleic acid structure from its sequence. Secondary structure can be predicted from one or several nucleic acid sequences. Tertiary structure can be predicted from the sequence, or by comparative modeling (when the structure of a homologous sequence is known). The problem of predicting nucleic acid secondary structure is dependent mainly on base pairing and base stacking interactions; many molecules have several possible three-dimensional structures, so predicting these structures remains out of reach unless obvious sequence and functional similarity to a known class of nucleic acid molecules, such as transfer RNA (tRNA) or microRNA (miRNA), is observed. Many secondary structure prediction methods rely on variations of dynamic programming and therefore are unable to efficiently identify pseudoknots. While the methods are similar, there are slight differences in the approach ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UC Berkeley
The University of California, Berkeley (UC Berkeley, Berkeley, Cal, or California) is a public university, public land-grant university, land-grant research university in Berkeley, California. Established in 1868 as the University of California, it is the state's first land-grant university and the founding campus of the University of California system. Its fourteen colleges and schools offer over 350 degree programs and enroll some 31,800 undergraduate and 13,200 graduate students. Berkeley ranks among the world's top universities. A founding member of the Association of American Universities, Berkeley hosts many leading research institutes dedicated to science, engineering, and mathematics. The university founded and maintains close relationships with three United States Department of Energy National Laboratories, national laboratories at Lawrence Berkeley National Laboratory, Berkeley, Lawrence Livermore National Laboratory, Livermore and Los Alamos National Laboratory, Los ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FASTA Format
In bioinformatics and biochemistry, the FASTA format is a text-based format for representing either nucleotide sequences or amino acid (protein) sequences, in which nucleotides or amino acids are represented using single-letter codes. The format also allows for sequence names and comments to precede the sequences. The format originates from the FASTA software package, but has now become a near universal standard in the field of bioinformatics. The simplicity of FASTA format makes it easy to manipulate and parse sequences using text-processing tools and scripting languages like the R programming language, Python, Ruby, Haskell, and Perl. Original format & overview The original FASTA/ Pearson format is described in the documentation for the FASTA suite of programs. It can be downloaded with any free distribution of FASTA (see fasta20.doc, fastaVN.doc or fastaVN.me—where VN is the Version Number). In the original format, a sequence was represented as a series of lines, each o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics Software
The list of bioinformatics software tools can be split up according to the license used: * List of proprietary bioinformatics software *List of open-source bioinformatics software Alternatively, here is a categorization according to the respective bioinformatics subfield specialized on: *Sequence analysis software **List of sequence alignment software **List of alignment visualization software **Alignment-free sequence analysis **De novo sequence assemblers **List of gene prediction software ** List of disorder prediction software ** List of Protein subcellular localization prediction tools ** List of phylogenetics software ** List of phylogenetic tree visualization software ** :Metagenomics_software *Structural biology software **List of molecular graphics systems **List of protein-ligand docking software ** List of RNA structure prediction software ** List of software for protein model error verification **List of protein secondary structure prediction programs **List of protein st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rfam
Rfam is a database containing information about non-coding RNA (ncRNA) families and other structured RNA elements. It is an annotated, open access database originally developed at the Wellcome Trust Sanger Institute in collaboration with Janelia Farm, and currently hosted at the European Bioinformatics Institute. Rfam is designed to be similar to the Pfam database for annotating protein families. Unlike proteins, ncRNAs often have similar secondary structure without sharing much similarity in the primary sequence. Rfam divides ncRNAs into families based on evolution from a common ancestor. Producing multiple sequence alignments (MSA) of these families can provide insight into their structure and function, similar to the case of protein families. These MSAs become more useful with the addition of secondary structure information. Rfam researchers also contribute to Wikipedia's RNA WikiProject. Uses The Rfam database can be used for a variety of functions. For each ncRNA f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Inside–outside Algorithm
For parsing algorithms in computer science, the inside–outside algorithm is a way of re-estimating production probabilities in a probabilistic context-free grammar. It was introduced by James K. Baker in 1979 as a generalization of the forward–backward algorithm for parameter estimation on hidden Markov models to stochastic context-free grammars. It is used to compute expectations, for example as part of the expectation–maximization algorithm In statistics, an expectation–maximization (EM) algorithm is an iterative method to find (local) maximum likelihood or maximum a posteriori (MAP) estimates of parameters in statistical models, where the model depends on unobserved latent variab ... (an unsupervised learning algorithm). Inside and outside probabilities The inside probability \beta_j(p,q) is the total probability of generating words w_p \cdots w_q, given the root nonterminal N^j and a grammar G: :\beta_j(p,q) = P(w_, N^j_, G) The outside probability \alpha_j(p,q) is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Smith–Waterman Algorithm
The Smith–Waterman algorithm performs local sequence alignment; that is, for determining similar regions between two strings of nucleic acid sequences or protein sequences. Instead of looking at the Needleman–Wunsch algorithm, entire sequence, the Smith–Waterman algorithm compares segments of all possible lengths and Mathematical optimization, optimizes the similarity measure. The algorithm was first proposed by Temple F. Smith and Michael S. Waterman in 1981. Like the Needleman–Wunsch algorithm, of which it is a variation, Smith–Waterman is a dynamic programming algorithm. As such, it has the desirable property that it is guaranteed to find the optimal local alignment with respect to the scoring system being used (which includes the substitution matrix and the Gap penalty, gap-scoring scheme). The main difference to the Needleman–Wunsch algorithm is that negative scoring matrix cells are set to zero, which renders the (thus positively scoring) local alignments visible ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Big O Notation
Big ''O'' notation is a mathematical notation that describes the limiting behavior of a function when the argument tends towards a particular value or infinity. Big O is a member of a family of notations invented by Paul Bachmann, Edmund Landau, and others, collectively called Bachmann–Landau notation or asymptotic notation. The letter O was chosen by Bachmann to stand for '' Ordnung'', meaning the order of approximation. In computer science, big O notation is used to classify algorithms according to how their run time or space requirements grow as the input size grows. In analytic number theory, big O notation is often used to express a bound on the difference between an arithmetical function and a better understood approximation; a famous example of such a difference is the remainder term in the prime number theorem. Big O notation is also used in many other fields to provide similar estimates. Big O notation characterizes functions according to their growth rates: d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dynamic Programming
Dynamic programming is both a mathematical optimization method and a computer programming method. The method was developed by Richard Bellman in the 1950s and has found applications in numerous fields, from aerospace engineering to economics. In both contexts it refers to simplifying a complicated problem by breaking it down into simpler sub-problems in a recursive manner. While some decision problems cannot be taken apart this way, decisions that span several points in time do often break apart recursively. Likewise, in computer science, if a problem can be solved optimally by breaking it into sub-problems and then recursively finding the optimal solutions to the sub-problems, then it is said to have '' optimal substructure''. If sub-problems can be nested recursively inside larger problems, so that dynamic programming methods are applicable, then there is a relation between the value of the larger problem and the values of the sub-problems.Cormen, T. H.; Leiserson, C. E.; Riv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Secondary Structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Types The most common secondary ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stockholm Format
Stockholm format is a multiple sequence alignment format used by Pfam, Rfam anDfam to disseminate protein, RNA and DNA sequence alignments. The alignment editorRalee,;()[aBb.-_--supports pseudoknot and further structure markup (see WUSS documentation) For protein [HGIEBTSCX] SA Surface Accessibility [0-9X] (0=0%-10%; ...; 9=90%-100%) TM TransMembrane [Mio] PP Posterior Probability [0-9*] (0=0.00-0.05; 1=0.05-0.15; *=0.95-1.00) LI LIgand binding AS Active Site pAS AS - Pfam predicted sAS AS - from SwissProt IN INtron (in or after) -2 For RNA tertiary interactions: ------------------------------ tWW WC/WC in trans For basepairs: AaBb...Zz">>AaBb...Zz For unpaired: cWH WC/Hoogsteen in cis cWS WC/SugarEdge in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stochastic Context-free Grammar
Grammar theory to model symbol strings originated from work in computational linguistics aiming to understand the structure of natural languages. Probabilistic context free grammars (PCFGs) have been applied in probabilistic modeling of RNA structures almost 40 years after they were introduced in computational linguistics. PCFGs extend context-free grammars similar to how hidden Markov models extend regular grammars. Each production is assigned a probability. The probability of a derivation (parse) is the product of the probabilities of the productions used in that derivation. These probabilities can be viewed as parameters of the model, and for large problems it is convenient to learn these parameters via machine learning. A probabilistic grammar's validity is constrained by context of its training dataset. PCFGs have application in areas as diverse as natural language processing to the study the structure of RNA molecules and design of programming languages. Designing efficient ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |