|
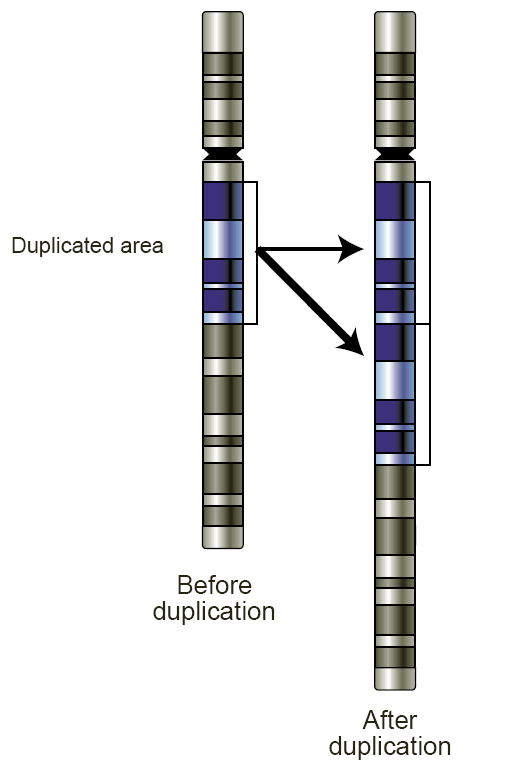
Segmental Duplication On The Human Y Chromosome
Segmental duplication are blocks of DNA ranging from 1 to 400 kb in length which recur at multiple sites within the genome, sharing greater than 90% similarity. Multiple studies have found a correlation between the location of segmental duplications and regions of chromosomal instability. This correlation suggests that they may be mediators of some genomic disorders. Segmental duplications are shown to be flanked on both sides by large homologous repeats, which exposes the region to recurrent rearrangement by nonallelic homologous recombination, leading to either deletion, duplication, or inversion of the original sequence. Background Finding segmental duplications Cataloging of segmental duplications was originally difficult due to its inconspicuousness, large size, and high degree of sequence similarity. This led to issues of interpreting separate loci as one sequence as these duplications are over-represented in unordered and unassigned contigs. Furthermore, these dup ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Segmental Duplication
Low copy repeats (LCRs), also known as segmental duplications (SDs), are highly homologous sequence elements within the eukaryotic genome. Repeats The repeats, or duplications, are typically 10–300 kb in length, and bear greater than 95% sequence identity. Though rare in most mammals, LCRs comprise a large portion of the human genome owing to a significant expansion during primate evolution. In humans, chromosomes Y and 22 have the greatest proportion of SDs: 50.4% and 11.9% respectively. Misalignment of LCRs during non-allelic homologous recombination (NAHR) is an important mechanism underlying the chromosomal microdeletion disorders as well as their reciprocal duplication partners. Many LCRs are concentrated in "hotspots", such as the 17p11-12 region, 27% of which is composed of LCR sequence. NAHR and non-homologous end joining Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence In Situ Hybridization
Fluorescence ''in situ'' hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only particular parts of a nucleic acid sequence with a high degree of sequence complementarity. It was developed by biomedical researchers in the early 1980s to detect and localize the presence or absence of specific DNA sequences on chromosomes. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling, medicine, and species identification. FISH can also be used to detect and localize specific RNA targets (mRNA, lncRNA and miRNA) in cells, circulating tumor cells, and tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression within cells and tissues. Probes – RNA and DNA In biology, a probe is a single strand of DNA or RNA that is complementary to a nucleotide sequence ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Copy-number Variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype. Copy number variations can be generally categorized into two main groups: short repeats and long repeats. However, there are no clear boundaries between the two groups and the classification depends on the nature of the loci of interest. Short repeats include mainly dinucleotide repeats (two repeating nucleotides e.g. A-C-A-C-A-C...) and trinucleotide repeats. Long r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-allelic Homologous Recombination
Non-allelic homologous recombination (NAHR) is a form of homologous recombination that occurs between two lengths of DNA that have high sequence similarity, but are not alleles. It usually occurs between sequences of DNA that have been previously duplicated through evolution, and therefore have low copy repeats (LCRs). These repeat elements typically range from 10–300 kb in length and share 95-97% sequence identity. During meiosis, LCRs can misalign and subsequent crossing-over can result in genetic rearrangement. When non-allelic homologous recombination occurs between different LCRs, deletions or further duplications of the DNA can occur. This can give rise to rare genetic disorders, caused by the loss or increased copy number of genes within the deleted or duplicated region. It can also contribute to the copy number variation seen in some gene clusters. As LCRs are often found in "hotspots" in the human genome, some chromosomal regions are particularly prone to NAHR. Rec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |