|
ScGET-seq
Single-cell genome and epigenome by transposases sequencing (scGET-seq) is a DNA sequencing method for profiling open and closed chromatin. In contrast to single-cell ATAC-seq, assay for transposase-accessible chromatin with sequencing (scATAC-seq), which only targets active euchromatin, scGET-seq is also capable of probing inactive heterochromatin. This is achieved through the use of TnH, which is created by linking the chromodomain (CD) of Heterochromatin protein 1, heterochromatin protein-1-alpha (HP-1\alpha) to the Tn5 transposase. TnH is then able to target histone 3 lysine 9 trimethylation (H3K9me3), a marker for heterochromatin. Akin to RNA velocity, which uses the ratio of RNA splicing, spliced to unspliced RNA to infer the kinetics of changes in gene expression over the course of cellular development, the ratio of TnH to Tn5 signals obtained from scGET-seq can be used to calculate chromatin velocity, which measures the dynamics of chromatin accessibility over the course ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATAC-seq
ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) is a laboratory technique used in molecular biology to assess genome-wide chromatin, chromatin accessibility. The technique was first described in 2013 as an alternative approach to MNase-seq, FAIRE-Seq and DNase-Seq but providing faster turnaround time, simplified protocol, and lower DNA input amount. Procedure ATAC-seq identifies accessible DNA regions by probing open chromatin with hyperactive mutant Transposase#Transposase Tn5, Tn5 Transposase that inserts sequencing adapters into open regions of the genome. While naturally occurring transposases have a low level of activity, ATAC-seq employs the mutated hyperactive transposase. In a process called "tagmentation", Tn5 transposase cleaves and tags double-stranded DNA with sequencing adaptors in a single enzymatic step. The tagged DNA fragments are then purified, Polymerase chain reaction, PCR-amplified, and sequenced using massive parallel sequencing, next-ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ScGET-seq Methods Overview
Single-cell genome and epigenome by transposases sequencing (scGET-seq) is a DNA sequencing method for profiling open and closed chromatin. In contrast to single-cell assay for transposase-accessible chromatin with sequencing (scATAC-seq), which only targets active euchromatin, scGET-seq is also capable of probing inactive heterochromatin. This is achieved through the use of TnH, which is created by linking the chromodomain (CD) of heterochromatin protein-1-alpha (HP-1\alpha) to the Tn5 transposase. TnH is then able to target histone 3 lysine 9 trimethylation (H3K9me3), a marker for heterochromatin. Akin to RNA velocity, which uses the ratio of spliced to unspliced RNA to infer the kinetics of changes in gene expression over the course of cellular development, the ratio of TnH to Tn5 signals obtained from scGET-seq can be used to calculate chromatin velocity, which measures the dynamics of chromatin accessibility over the course of cellular developmental pathways. History T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, Genographic Project, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid advancements in DNA seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metastasis
Metastasis is a pathogenic agent's spreading from an initial or primary site to a different or secondary site within the host's body; the term is typically used when referring to metastasis by a cancerous tumor. The newly pathological sites, then, are metastases (mets). It is generally distinguished from cancer invasion, which is the direct extension and penetration by cancer cells into neighboring tissues. Cancer occurs after cells are genetically altered to proliferate rapidly and indefinitely. This uncontrolled proliferation by mitosis produces a primary tumor, primary tumour heterogeneity, heterogeneic tumour. The cells which constitute the tumor eventually undergo metaplasia, followed by dysplasia then anaplasia, resulting in a Malignancy, malignant phenotype. This malignancy allows for invasion into the circulation, followed by invasion to a second site for tumorigenesis. Some cancer cells, known as circulating tumor cells (CTCs), are able to penetrate the walls of lymp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Matrix Factorization
In the mathematical discipline of linear algebra, a matrix decomposition or matrix factorization is a factorization of a matrix into a product of matrices. There are many different matrix decompositions; each finds use among a particular class of problems. Example In numerical analysis, different decompositions are used to implement efficient matrix algorithms. For example, when solving a system of linear equations A \mathbf = \mathbf, the matrix ''A'' can be decomposed via the LU decomposition. The LU decomposition factorizes a matrix into a lower triangular matrix ''L'' and an upper triangular matrix ''U''. The systems L(U \mathbf) = \mathbf and U \mathbf = L^ \mathbf require fewer additions and multiplications to solve, compared with the original system A \mathbf = \mathbf, though one might require significantly more digits in inexact arithmetic such as floating point. Similarly, the QR decomposition expresses ''A'' as ''QR'' with ''Q'' an orthogonal matrix and ''R'' an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
K-nearest Neighbors Algorithm
In statistics, the ''k''-nearest neighbors algorithm (''k''-NN) is a Non-parametric statistics, non-parametric supervised learning method. It was first developed by Evelyn Fix and Joseph Lawson Hodges Jr., Joseph Hodges in 1951, and later expanded by Thomas M. Cover, Thomas Cover. Most often, it is used for statistical classification, classification, as a ''k''-NN classifier, the output of which is a class membership. An object is classified by a plurality vote of its neighbors, with the object being assigned to the class most common among its ''k'' nearest neighbors (''k'' is a positive integer, typically small). If ''k'' = 1, then the object is simply assigned to the class of that single nearest neighbor. The ''k''-NN algorithm can also be generalized for regression analysis, regression. In ''-NN regression'', also known as ''nearest neighbor smoothing'', the output is the property value for the object. This value is the average of the values of ''k'' nearest neighbo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Principal Component Analysis
Principal component analysis (PCA) is a linear dimensionality reduction technique with applications in exploratory data analysis, visualization and data preprocessing. The data is linearly transformed onto a new coordinate system such that the directions (principal components) capturing the largest variation in the data can be easily identified. The principal components of a collection of points in a real coordinate space are a sequence of p unit vectors, where the i-th vector is the direction of a line that best fits the data while being orthogonal to the first i-1 vectors. Here, a best-fitting line is defined as one that minimizes the average squared perpendicular distance from the points to the line. These directions (i.e., principal components) constitute an orthonormal basis in which different individual dimensions of the data are linearly uncorrelated. Many studies use the first two principal components in order to plot the data in two dimensions and to visually identi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reference Genome
A reference genome (also known as a reference assembly) is a digital nucleic acid sequence database, assembled by scientists as a representative example of the genome, set of genes in one idealized individual organism of a species. As they are assembled from the sequencing of DNA from a number of individual donors, reference genomes do not accurately represent the set of genes of any single individual organism. Instead, a reference provides a haploid mosaic of different DNA sequences from each donor. For example, one of the most recent human reference genomes, assembly ''GRCh38, GRCh38/hg38'', is derived from >60 genomic library, genomic clone libraries. There are reference genomes for multiple species of viruses, bacteria, fungus, plants, and animals. Reference genomes are typically used as a guide on which new genomes are built, enabling them to be assembled much more quickly and cheaply than the initial Human Genome Project. Reference genomes can be accessed online at several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, a Guanine, G nucleotide present at a specific location in a reference genome may be replaced by an Adenine, A in a minority of individuals. The two possible nucleotide variations of this SNP – G or A – are called alleles. SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the Factor H, CFH gene is associated with increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs. For example, two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
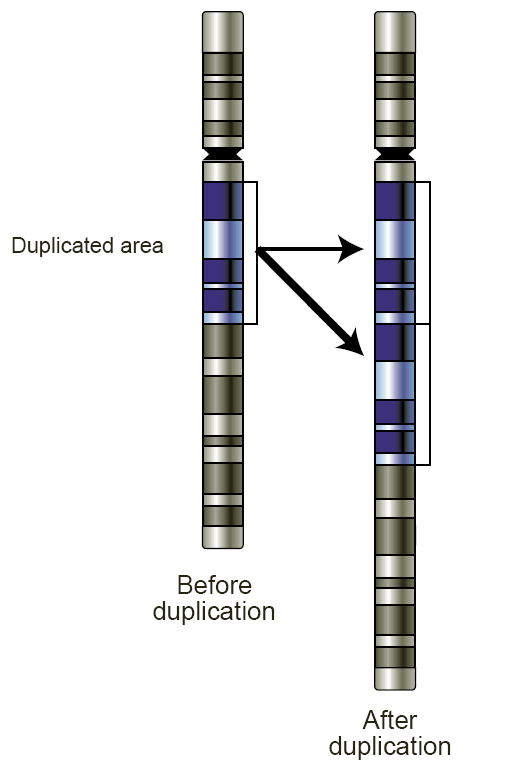
Copy Number Variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype. Copy number variations can be generally categorized into two main groups: short repeats and long repeats. However, there are no clear boundaries between the two groups and the classification depends on the nature of the loci of interest. Short repeats include mainly dinucleotide repeats (two repeating nucleotides e.g. A-C-A-C-A-C...) and trinucleotide repeats. Lon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins Histone H2A, H2A, Histone H2B, H2B, Histone H3, H3, and Histone H4, H4. DNA must be compacted into nucleosomes to fit within the cell nucleus. In addition to nucleosome wrapping, eukaryotic chromatin is further compacted by being folded into a series of more complex structures, eventually forming a chromosome. Each human cell contains about 30 million nucleosomes. Nucleosomes are thought to carry Epigenetics, epigenetically inherited information in the form of coval ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |