|
SUPERFAMILY
SUPERFAMILY is a database and search platform of structural and functional annotation for all proteins and genomes. It classifies amino acid sequences into known structural domains, especially into SCOP superfamilies. Domains are functional, structural, and evolutionary units that form proteins. Domains of common Ancestry are grouped into superfamilies. The domains and domain superfamilies are defined and described in SCOP. Superfamilies are groups of proteins which have structural evidence to support a common evolutionary ancestor but may not have detectable sequence homology. Annotations The SUPERFAMILY annotation is based on a collection of hidden Markov models (HMM), which represent structural protein domains at the SCOP superfamily level. A superfamily groups together domains which have an evolutionary relationship. The annotation is produced by scanning protein sequences from completely sequenced genomes against the hidden Markov models. For each protein you can: * S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or Disulfide bond, disulfide bridges. Domains often form functional units, such as the calcium-binding EF-hand, EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimera (protein), chimeric ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Classification Of Proteins
The Structural Classification of Proteins (SCOP) database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine the evolutionary relationship between proteins. Proteins with the same shapes but having little sequence or functional similarity are placed in different superfamilies, and are assumed to have only a very distant common ancestor. Proteins having the same shape and some similarity of sequence and/or function are placed in "families", and are assumed to have a closer common ancestor. Similar to CATH and Pfam databases, SCOP provides a classification of individual structural domains of proteins, rather than a classification of the entire proteins which may include a significant number of different domains. The SCOP database is freely accessible on the internet. SCOP was created in 1994 in the Centre for Protein Engineering and the Lab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Superfamily
A protein superfamily is the largest grouping (clade) of proteins for which common ancestry can be inferred (see homology (biology), homology). Usually this common ancestry is inferred from structural alignment and mechanistic similarity, even if no sequence similarity is evident. Sequence homology can then be deduced even if not apparent (due to low sequence similarity). Superfamilies typically contain several protein families which show sequence similarity within each family. The term ''protein clan'' is commonly used for protease and glycosyl hydrolases superfamilies based on the MEROPS and CAZy classification systems. Identification Superfamilies of proteins are identified using a number of methods. Closely related members can be identified by different methods to those needed to group the most evolutionarily divergent members. Sequence similarity Historically, the similarity of different amino acid sequences has been the most common method of inferring Sequence homology, h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Julian Gough (scientist)
Julian John Thurstan Gough (born 1974) was a Group Leader in the Laboratory of Molecular Biology (LMB) of the Medical Research Council (MRC). He was previously a professor of bioinformatics at the University of Bristol. Education Gough was educated at The Perse School in Cambridge and the University of Bristol where he was awarded a joint honours degree in Mathematics and Physics in 1998. He went on to complete his PhD in the Laboratory of Molecular Biology (LMB) supervised by Cyrus Chothia on genome analysis and protein structure as a postgraduate student of Sidney Sussex College, Cambridge, graduating in 2001. Career and research Following his PhD, Gough completed postdoctoral research at the LMB and Stanford University, with Michael Levitt. Subsequently, he was a scientist at RIKEN in Tokyo, a Professor member of faculty in the Computer Science department at the University of Bristol, where he worked from 2007-2017, and then a programme leader at the MRC Laboratory of Mol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyrus Chothia
Cyrus Homi Chothia (19 February 1942 – 26 November 2019) was an English biochemist who was an emeritus scientist at the Medical Research Council (MRC) Laboratory of Molecular Biology (LMB) at the University of Cambridge and emeritus fellow of Wolfson College, Cambridge. Education Chothia was educated at Alleyn's School, then went to study at Durham University graduating with a Bachelor of Science degree in 1965. Chothia then completed a Master of Science degree at Birkbeck College in 1967 and a PhD from University College London under the supervision of , the son of Linus Pauling. Research and career After his PhD Chothia worked in the Laboratory of Molecular Biology (LMB) for three years. He then worked with Michael Levitt at the Weizmann Institute of Science followed by two years with Joel Janin at the Institut Pasteur in Paris. In 1976 Chothia returned to England to work at University College London and the LMB. With Arthur Lesk he showed that proteins adapt to mutatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Tree
A phylogenetic tree or phylogeny is a graphical representation which shows the evolutionary history between a set of species or taxa during a specific time.Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA. In other words, it is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. In evolutionary biology, all life on Earth is theoretically part of a single phylogenetic tree, indicating common ancestry. Phylogenetics is the study of phylogenetic trees. The main challenge is to find a phylogenetic tree representing optimal evolutionary ancestry between a set of species or taxa. Computational phylogenetics (also phylogeny inference) focuses on the algorithms involved in finding optimal phylogenetic tree in the phylogenetic landscape. Phylogenetic trees may be rooted or unrooted. In a ''rooted'' p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
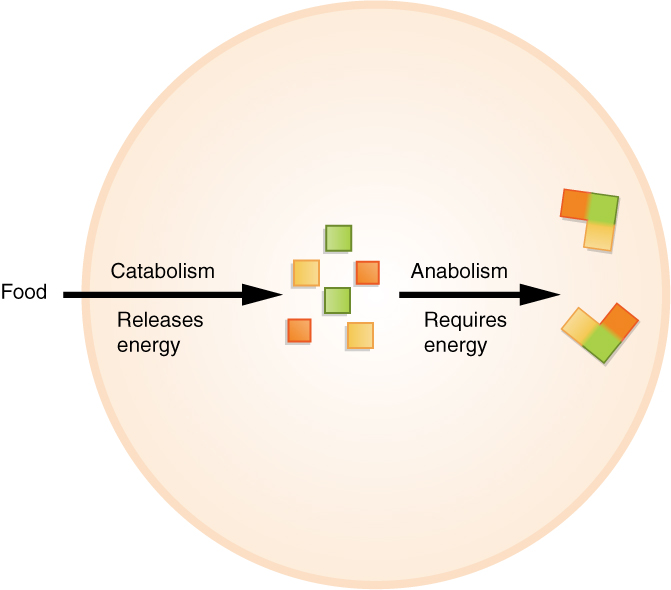
Metabolism
Metabolism (, from ''metabolē'', "change") is the set of life-sustaining chemical reactions in organisms. The three main functions of metabolism are: the conversion of the energy in food to energy available to run cellular processes; the conversion of food to building blocks of proteins, lipids, nucleic acids, and some carbohydrates; and the elimination of metabolic wastes. These enzyme-catalyzed reactions allow organisms to grow and reproduce, maintain their Structures#Biological, structures, and respond to their environments. The word ''metabolism'' can also refer to the sum of all chemical reactions that occur in living organisms, including digestion and the transportation of substances into and between different cells, in which case the above described set of reactions within the cells is called intermediary (or intermediate) metabolism. Metabolic reactions may be categorized as ''catabolic''—the ''breaking down'' of compounds (for example, of glucose to pyruvate by c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Anabolism
Anabolism () is the set of metabolic pathways that construct macromolecules like DNA or RNA from smaller units. These reactions require energy, known also as an Endergonic reaction, endergonic process. Anabolism is the building-up aspect of metabolism, whereas catabolism is the breaking-down aspect. Anabolism is usually synonymous with biosynthesis. Pathway Polymerization, an anabolic pathway used to build macromolecules such as nucleic acids, proteins, and polysaccharides, uses condensation reactions to join monomers. Macromolecules are created from smaller molecules using enzymes and Cofactor (biochemistry), cofactors. Energy source Anabolism is powered by catabolism, where large molecules are broken down into smaller parts and then used up in cellular respiration. Many anabolic processes are powered by the ATP hydrolysis, cleavage of adenosine triphosphate (ATP). Anabolism usually involves Redox, reduction and decreases entropy, making it unfavorable without energy input. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). This structural data is obtained and deposited by biologists and biochemists worldwide through the use of experimental methodologies such as X-ray crystallography, Nuclear magnetic resonance spectroscopy of proteins, NMR spectroscopy, and, increasingly, cryo-electron microscopy. All submitted data are reviewed by expert Biocuration, biocurators and, once approved, are made freely available on the Internet under the CC0 Public Domain Dedication. Global access to the data is provided by the websites of the wwPDB member organizations (PDBe, PDBj, RCSB PDB, and BMRB). The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Transduction
Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a biochemical cascade, series of molecular events. Proteins responsible for detecting stimuli are generally termed receptor (biology), receptors, although in some cases the term sensor is used. The changes elicited by ligand (biochemistry), ligand binding (or signal sensing) in a receptor give rise to a biochemical cascade, which is a chain of biochemical events known as a Cell signaling#Signaling pathways, signaling pathway. When signaling pathways interact with one another they form networks, which allow cellular responses to be coordinated, often by combinatorial signaling events. At the molecular level, such responses include changes in the transcription (biology), transcription or translation (biology), translation of genes, and post-translational modification, post-translational and conformational changes in proteins, as well as changes in their location. These molecula ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (biology)
In biology, translation is the process in living Cell (biology), cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotides in the RNA. The nucleotides are considered three at a time. Each such triple results in the addition of one specific amino acid to the protein being generated. The matching from nucleotide triple to amino acid is called the genetic code. The translation is performed by a large complex of functional RNA and proteins called ribosomes. The entire process is called gene expression. In translation, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later protein folding, folds into an Activation energy, active protein and performs its functions in the cell. The polypeptide can also start folding during protein synthesis. The ribosome facilitates decoding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |