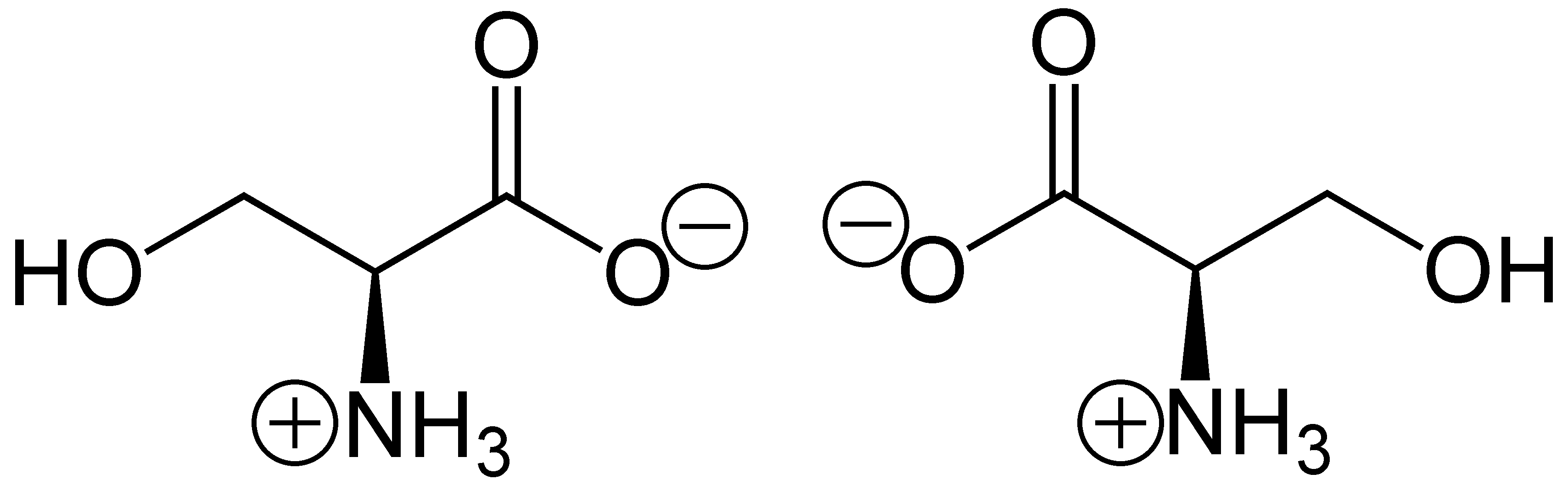|
SNX8
The SNX8 is a sorting nexin protein involved in intracellular molecular traffic from the early endosomes to the TGN. It is suggested that it acts as an adaptor protein in events related to immune response and cholesterol regulation, for example. As a protein of the SNXs family, the SNX8 is formed of 465 aminoacids and presents a BAR-domain and a PX-domain which are very relevant in relation to its functions. Furthermore, SNX8 study is motivated by its medical significance in relation to diseases such as Alzheimer's Disease, cancer, neurodevelopmental malformations and to its role in fighting against viral infections. Structure Sorting nexins (SNXs) SNX8 belongs to the sorting nexin family of proteins, which mainly contain two functional membrane-binding that allow SNXs to have different roles in endosomal sorting and protein trafficking thanks to its membrane curvature ability. To begin with, SNX-PX is a distinct phosphoinositide (PI)-binding domain. The preferential interacti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sorting Nexin
Sorting nexins are a large group of proteins that are localized in the cytoplasm and have the potential for membrane association either through their lipid-binding PX domain (a phospholipid-binding motif) or through protein–protein interactions with membrane-associated protein complexes Some members of this family have been shown to facilitate protein sorting. Family members In humans, sorting nexins are transcribed from the following genes: Structure Sorting nexins either consist solely of a PX domain (e.g. SNX3) or have a modular structure made up of the PX and additional domains. A subgroup of sorting nexins (comprising, in humans, SNX1, SNX2, SNX4, SNX5, SNX6, SNX7, SNX8, SNX9, SNX18, SNX30, SNX32 and SNX33) possess a BAR domain at their C-terminus. (The BAR domain of SNXs 1, 2, 4, 7, 8 and 30 is classified by pfam as 'Vps5 C terminal like'.) An example of a sorting nexin domain structure can be seen here for SNX1: # NTD – N-terminal domain # PX dom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bar Domain
In molecular biology, BAR domains are highly conserved protein dimerisation domains that occur in many proteins involved in membrane dynamics in a cell. The BAR domain is banana-shaped and binds to membrane via its concave face. It is capable of sensing membrane curvature by binding preferentially to curved membranes. BAR domains are named after three proteins that they are found in: Bin, Amphiphysin and Rvs. BAR domains occur in combinations with other domains Many BAR family proteins contain alternative lipid specificity domains that help target these protein to particular membrane compartments. Some also have SH3 domains that bind to dynamin and thus proteins like amphiphysin and endophilin are implicated in the orchestration of vesicle scission. N-BAR domain Some BAR domain containing proteins have an N-terminal amphipathic helix preceding the BAR domain. This helix inserts (like in the epsin ENTH domain) into the membrane and induces curvature, which is stabilise ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PX Domain
The PX domain is a phosphoinositide-binding structural domain involved in targeting of proteins to cell membranes. This domain was first found in P40phox and p47phox domains of NADPH oxidase (phox stands for phagocytic oxidase). It was also identified in many other proteins involved in membrane trafficking, including nexins, Phospholipase D, and phosphoinositide-3-kinases. The PX domain is structurally conserved in eukaryotes, although amino acid sequences show little similarity. PX domains interact primarily with PtdIns(3)P lipids. However some of them bind to phosphatidic acid, PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3. The PX-domain can also interact with other domains and proteins. Human proteins containing this domain Sorting nexins contain this domain. Other examples include: * HS1BP3 * KIF16B (SNX23) * NCF1; NCF1C; NCF4; NISCH * PIK3C2A; PIK3C2B; PIK3C2G; PLD1; PLD2; PXK * RPS6KC1 * SGK3 Serine/threonine-protein kinase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Mass
The molecular mass (''m'') is the mass of a given molecule: it is measured in daltons (Da or u). Different molecules of the same compound may have different molecular masses because they contain different isotopes of an element. The related quantity relative molecular mass, as defined by IUPAC, is the ratio of the mass of a molecule to the unified atomic mass unit (also known as the dalton) and is unitless. The molecular mass and relative molecular mass are distinct from but related to the molar mass. The molar mass is defined as the mass of a given substance divided by the amount of a substance and is expressed in g/mol. That makes the molar mass an average of many particles or molecules, and the molecular mass the mass of one specific particle or molecule. The molar mass is usually the more appropriate figure when dealing with macroscopic (weigh-able) quantities of a substance. The definition of molecular weight is most authoritatively synonymous with relative molecular mass; ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysosome
A lysosome () is a membrane-bound organelle found in many animal cells. They are spherical vesicles that contain hydrolytic enzymes that can break down many kinds of biomolecules. A lysosome has a specific composition, of both its membrane proteins, and its lumenal proteins. The lumen's pH (~4.5–5.0) is optimal for the enzymes involved in hydrolysis, analogous to the activity of the stomach. Besides degradation of polymers, the lysosome is involved in various cell processes, including secretion, plasma membrane repair, apoptosis, cell signaling, and energy metabolism. Lysosomes act as the waste disposal system of the cell by digesting used materials in the cytoplasm, from both inside and outside the cell. Material from outside the cell is taken up through endocytosis, while material from the inside of the cell is digested through autophagy. The sizes of the organelles vary greatly—the larger ones can be more than 10 times the size of the smaller ones. They we ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Vacuole
A vacuole () is a membrane-bound organelle which is present in plant and fungal cells and some protist, animal, and bacterial cells. Vacuoles are essentially enclosed compartments which are filled with water containing inorganic and organic molecules including enzymes in solution, though in certain cases they may contain solids which have been engulfed. Vacuoles are formed by the fusion of multiple membrane vesicles and are effectively just larger forms of these. The organelle has no basic shape or size; its structure varies according to the requirements of the cell. Discovery Contractile vacuoles ("stars") were first observed by Spallanzani (1776) in protozoa, although mistaken for respiratory organs. Dujardin (1841) named these "stars" as ''vacuoles''. In 1842, Schleiden applied the term for plant cells, to distinguish the structure with cell sap from the rest of the protoplasm. In 1885, de Vries named the vacuole membrane as tonoplast. Function The function and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Homology
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are estimated to constitute 1% of all described fungal species. Yeasts are unicellular organisms that evolved from multicellular ancestors, with some species having the ability to develop multicellular characteristics by forming strings of connected budding cells known as pseudohyphae or false hyphae. Yeast sizes vary greatly, depending on species and environment, typically measuring 3–4 µm in diameter, although some yeasts can grow to 40 µm in size. Most yeasts reproduce asexually by mitosis, and many do so by the asymmetric division process known as budding. With their single-celled growth habit, yeasts can be contrasted with molds, which grow hyphae. Fungal species that can take both forms (depending on temperature or other conditions) are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Phosphorylation
Protein phosphorylation is a reversible post-translational modification of proteins in which an amino acid residue is phosphorylated by a protein kinase by the addition of a covalently bound phosphate group. Phosphorylation alters the structural conformation of a protein, causing it to become either activated or deactivated, or otherwise modifying its function. Approximately 13000 human proteins have sites that are phosphorylated. The reverse reaction of phosphorylation is called dephosphorylation, and is catalyzed by protein phosphatases. Protein kinases and phosphatases work independently and in a balance to regulate the function of proteins. The amino acids most commonly phosphorylated are serine, threonine, tyrosine in eukaryotes, and also histidine in prokaryotes and plants (though it is now known to be common in humans). These phosphorylations play important and well-characterized roles in signaling pathways and metabolism. However, other amino acids can also be phospho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L- stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |