|
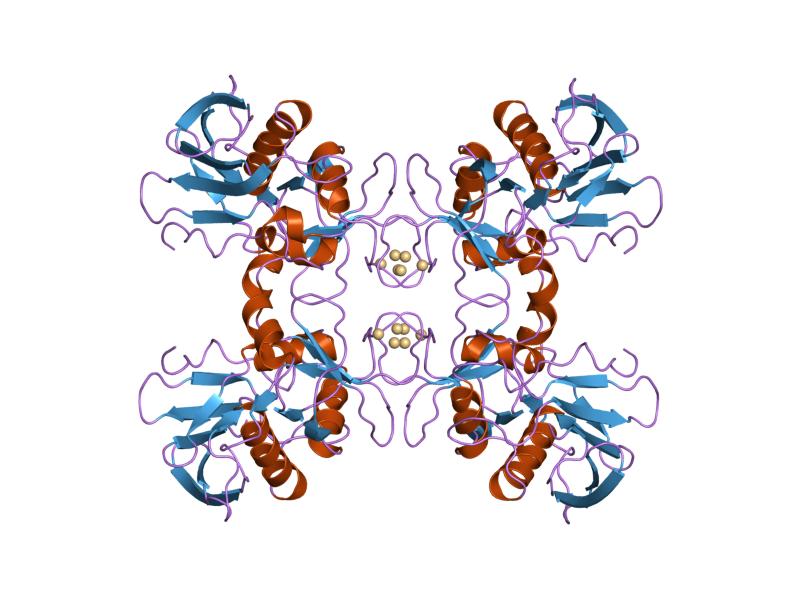
RopB
RopB transcriptional regulator, also known as RopB/Rgg transcriptional regulator is a transcriptional regulator protein that regulates expression of the extracellularly secreted cysteine protease streptococcal pyrogenic exotoxin B (speB or streptopain), which is an important virulence factor of ''Streptococcus pyogenes'' and is responsible for the dissemination of a host of infectious diseases including strep throat, impetigo, streptococcal toxic shock syndrome, necrotizing fasciitis, and scarlet fever. Functional studies suggest that the ropB multigene regulon is responsible for not only global regulation of virulence but also a wide range of functions from stress response, metabolic function, and two-component signaling. Structural studies implicate ropB's regulatory action being reliant on a complex interaction involving quorum sensing with the leaderless peptide signal speB-inducing peptide (SIP); acting in conjunction with a pH sensitive histidine switch. Discovery O ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Streptococcus Pyogenes
''Streptococcus pyogenes'' is a species of Gram-positive, aerotolerant bacteria in the genus '' Streptococcus''. These bacteria are extracellular, and made up of non-motile and non-sporing cocci (round cells) that tend to link in chains. They are clinically important for humans, as they are an infrequent, but usually pathogenic, part of the skin microbiota that can cause group A streptococcal infection. ''S. pyogenes'' is the predominant species harboring the Lancefield group A antigen, and is often called group A ''Streptococcus'' (GAS). However, both '' Streptococcus dysgalactiae'' and the '' Streptococcus anginosus'' group can possess group A antigen as well. Group A streptococci, when grown on blood agar, typically produce small (2–3 mm) zones of beta-hemolysis, a complete destruction of red blood cells. The name group A (beta-hemolytic) ''Streptococcus'' is thus also used. The species name is derived from Greek words meaning 'a chain' () of berries ( atiniz ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Streptococcal Pyrogenic Exotoxin
Streptococcal pyrogenic exotoxins also known as erythrogenic toxins, are exotoxins secreted by strains of the bacterial species ''Streptococcus pyogenes''. SpeA and speC are superantigens, which induce inflammation by nonspecifically activating T cells and stimulating the production of inflammatory cytokines. SpeB, the most abundant streptococcal extracellular protein, is a cysteine protease. Pyrogenic exotoxins are implicated as the causative agent of scarlet fever and streptococcal toxic shock syndrome. There is no consensus on the exact number of pyrogenic exotoxins. Serotypes A, B, and C are the most extensively studied and recognized by all sources, but others note up to thirteen distinct types, categorizing speF through speM as additional superantigens. Erythrogenic toxins are known to damage the plasma membranes of blood capillaries under the skin and produce a red skin rash (characteristic of scarlet fever). Past studies have shown that multiple variants of erythrogenic toxin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Streptopain
Streptopain, also known as streptococcal pyrogenic exotoxin B (SpeB) is a streptococcal cysteine protease. Other names include Streptococcus peptidase A, Streptococcus protease, and streptococcal cysteine proteinase. Streptopain catalyses the following chemical reaction : Preferential cleavage with hydrophobic residues at P2, P1 and P1' It is isolated from the group A bacterium ''Streptococcus pyogenes'' and acts as a virulence factor Virulence factors (preferably known as pathogenicity factors or effectors in botany) are cellular structures, molecules and regulatory systems that enable microbial pathogens (bacteria, viruses, fungi, and protozoa) to achieve the following: * c .... See also * RopB transcriptional regulator References External links * {{Portal bar, Biology, border=no EC 3.4.22 Streptococcal proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Palindromic Sequence
A palindromic sequence is a nucleic acid sequence in a double-stranded DNA or RNA molecule whereby reading in a certain direction (e.g. 5' to 3') on one strand is identical to the sequence in the same direction (e.g. 5' to 3') on the complementary strand. This definition of palindrome thus depends on complementary strands being palindromic of each other. The meaning of palindrome in the context of genetics is slightly different from the definition used for words and sentences. Since a double helix is formed by two paired antiparallel strands of nucleotides that run in opposite directions, and the nucleotides always pair in the same way (adenine (A) with thymine (T) in DNA or uracil (U) in RNA; cytosine (C) with guanine (G)), a (single-stranded) nucleotide sequence is said to be a palindrome if it is equal to its reverse complement. For example, the DNA sequence ACCTAGGT is palindromic with its nucleotide-by-nucleotide complement TGGATCCA because reversing the order of the n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparison Between Rgg And RopB Chromosomal Regions
Comparison or comparing is the act of evaluating two or more things by determining the relevant, comparable characteristics of each thing, and then determining which characteristics of each are similar to the other, which are different, and to what degree. Where characteristics are different, the differences may then be evaluated to determine which thing is best suited for a particular purpose. The description of similarities and differences found between the two things is also called a comparison. Comparison can take many distinct forms, varying by field: To compare things, they must have characteristics that are similar enough in relevant ways to merit comparison. If two things are too different to compare in a useful way, an attempt to compare them is colloquially referred to in English as "comparing apples and oranges." Comparison is widely used in society, in science and the arts. General usage Comparison is a natural activity, which even animals engage in when decidin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Overview For transcription to take place, the enzyme that synthesizes RNA, known as RNA polymerase, must attach to the DNA near a gene. Promoters contain specific DNA sequences such as response elements that provide a secure initial binding site for RNA polymerase and for proteins called transcription factors that recruit RNA polymerase. These transcription factor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Consensus Sequence
In molecular biology and bioinformatics, the consensus sequence (or canonical sequence) is the calculated sequence of most frequent residues, either nucleotide or amino acid, found at each position in a sequence alignment. It represents the results of multiple sequence alignments in which related sequences are compared to each other and similar sequence motifs are calculated. Such information is important when considering sequence-dependent enzymes such as RNA polymerase.Pierce, Benjamin A. 2002. Genetics : A Conceptual Approach. 1st ed. New York: W.H. Freeman and Co. Biological significance A protein binding site, represented by a consensus sequence, may be a short sequence of nucleotides which is found several times in the genome and is thought to play the same role in its different locations. For example, many transcription factors recognize particular patterns in the promoters of the genes they regulate. In the same way, restriction enzymes usually have palindromic consensu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polypyrimidine Tract
{{Short description, A pyrimidine-rich sequence involved in pre-messenger RNA maturation The polypyrimidine tract is a region of pre-messenger RNA (mRNA) that promotes the assembly of the spliceosome, the protein complex specialized for carrying out RNA splicing during the process of post-transcriptional modification. The region is rich with pyrimidine nucleotides, especially uracil, and is usually 15–20 base pairs long, located about 5–40 base pairs before the 3' end of the intron to be spliced.Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipursky SL, Darnell J. (2004). ''Molecular Cell Biology.'' WH Freeman: New York, NY. 5th ed. A number of protein factors bind to or associate with the polypyrimidine tract, including the spliceosome component U2AF and the '' polypyrimidine tract-binding protein'' (PTB), which plays a regulatory role in alternative splicing. PTB's primary function is in exon silencing, by which a particular exon region normally spliced i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcriptional Regulator
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA ( transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology. The regulation of transcription is a vital process in all living organisms. It is orchestrated by transcription factors and other proteins working in concert to finely tune th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intrinsic Termination
Intrinsic, or rho-independent termination, is a process to signal the end of transcription and release the newly constructed RNA molecule. In bacteria such as ''E. coli'', transcription is terminated either by a rho-dependent process or rho-independent process. In the Rho-dependent process, the rho-protein locates and binds the signal sequence in the mRNA and signals for cleavage. Contrarily, intrinsic termination does not require a special protein to signal for termination and is controlled by the specific sequences of RNA. When the termination process begins, the transcribed mRNA forms a stable secondary structure hairpin loop, also known as a stem-loop. This RNA hairpin is followed by multiple uracil nucleotides. The bonds between uracil (rU) and adenine (dA) are very weak. A protein bound to RNA polymerase (nusA) binds to the stem-loop structure tightly enough to cause the polymerase to temporarily stall. This pausing of the polymerase coincides with transcription of the poly- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalysis, catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks Covalent bond, bonds. Proteases are involved in numerous biological pathways, including Digestion#Protein digestion, digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently convergent evolution, evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Classification Based on catalytic residue Proteases can be classified into seven broad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |