|
Richard A. Rachubinski
Richard A. Rachubinski is a Canadian cell biologist and academic. He serves as a Professor at the University of Alberta in the Department of Cell Biology. Rachubinski's research centers on understanding how Peroxisome, peroxisomes—essential Organelle, organelles involved in lipid metabolism, redox balance, and cellular detoxification—are assembled, maintained, and inherited during cell division. Using model organisms such as ''Yarrowia lipolytica'', ''Saccharomyces cerevisiae'', and ''Drosophila melanogaster'', his lab has uncovered fundamental genetic and molecular pathways that govern peroxisome biogenesis and inheritance. A major focus of Rachubinski's work has been elucidating the mechanisms of peroxisomal protein targeting and import. He was among the first to demonstrate that peroxisomes can import fully folded, and even Oligomer, oligomeric, protein complexes—including dimers like thiolase—challenging the prevailing dogma that proteins must unfold to cross organell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactions. Though cells and other microscopic structures had been observed in living organisms as early as the 18th century, a detailed understanding of the mechanisms and interactions governing their behavior did not emerge until the 20th century, when technologies used in physics and chemistry had advanced sufficiently to permit their application in the biological sciences. The term 'molecular biology' was first used in 1945 by the English physicist William Astbury, who described it as an approach focused on discerning the underpinnings of biological phenomena—i.e. uncovering the physical and chemical structures and properties of biological molecules, as well as their interactions with other molecules and how these interactions explain observ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saccharomyces Cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like '' Escherichia coli'' as the model bacterium. It is the microorganism which causes many common types of fermentation. ''S. cerevisiae'' cells are round to ovoid, 5–10 μm in diameter. It reproduces by budding. Many proteins important in human biology were first discovered by studying their homologs in yeast; these proteins include cell cycle proteins, signaling proteins, and protein-processing enzymes. ''S. cerevisiae'' is currently the only yeast cell known to have Berkeley bodies present, which are involved in particular secretory pathways. Antibodies again ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Immunometabolism
Immunometabolism is a branch of biology that studies the interplay between metabolism and immunology in all organisms. In particular, immunometabolism is the study of the molecular and biochemical underpinnings for i) the metabolic regulation of immune function, and ii) the regulation of metabolism by molecules and cells of the immune system. Further categorization includes i) systemic immunometabolism and ii) cellular immunometabolism. Immunometabolism includes metabolic inflammation:a chronic, systemic, low grade inflammation, orchestrated by metabolic deregulation caused by obesity or aging. Immunometabolism first appears in academic literature in 2011, where it is defined as "an emerging field of investigation at the interface between the historically distinct disciplines of immunology and metabolism." A later article defines immunometabolism as describing "the changes that occur in intracellular metabolic pathways in immune cells during activation". Broadly, immunometabolic re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peroxisome Biogenesis Disorders
Peroxisomal disorders represent a class of medical conditions caused by defects in peroxisome functions. This may be due to defects in single enzymes important for peroxisome function or in peroxins, proteins encoded by ''PEX'' genes that are critical for normal peroxisome assembly and biogenesis. Peroxisome biogenesis disorders Peroxisome biogenesis disorders (PBDs) include the Zellweger syndrome spectrum (PBD-ZSD) and rhizomelic chondrodysplasia punctata type 1 (RCDP1). PBD-ZSD represents a continuum of disorders including infantile Refsum disease, neonatal adrenoleukodystrophy, and Zellweger syndrome. Collectively, PBDs are autosomal recessive developmental brain disorders that also result in skeletal and craniofacial dysmorphism, liver dysfunction, progressive sensorineural hearing loss, and retinopathy. PBD-ZSD is most commonly caused by mutations in the ''PEX1'', ''PEX6'', ''PEX10'', ''PEX12'', and ''PEX26'' genes. This results in the over-accumulation of very long chain ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peroxin
Peroxins (or peroxisomal/peroxisome biogenesis factors) represent several protein families found in peroxisomes. Deficiencies are associated with several peroxisomal disorders. Peroxins serve several functions including the recognition of cytoplasmic proteins that contain peroxisomal targeting signals (PTS) that tag them for transport by peroxisomal proteins to the peroxisome. Peroxins are structurally diverse and have been classified to different protein families. Some of them were predicted to be single-pass transmembrane proteins, for example Peroxisomal biogenesis factor 11. Genes * PEX1 * PEX2 * PEX3 * PEX5 Peroxisomal targeting signal 1 receptor (PTS1R) is a protein that in humans is encoded by the ''PEX5'' gene. PTS1R is a peroxisomal targeting sequence involved in the specific transport of molecules for oxidation inside the peroxisome. SKL bin ... * PEX6 * PEX7 * PEX10 * PEX11A, PEX11B, PEX11G * PEX12 * PEX13 * PEX14 * PEX16 * PEX19 * PEX26 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peroxisome-proliferating
In the field of molecular biology, the peroxisome proliferator–activated receptors (PPARs) are a group of nuclear receptor proteins that function as transcription factors regulating gene expression. PPARs play essential roles in regulating cellular differentiation, development, and metabolism (carbohydrate, lipid, protein), and tumorigenesis Nomenclature and tissue distribution Three types of PPARs have been identified: alpha, gamma, and delta (beta): * α (alpha) - expressed in liver, kidney, heart, muscle, adipose tissue, and others * β/δ (beta/delta) - expressed in many tissues, especially in brain, adipose tissue, and skin * γ (gamma) - although transcribed by the same gene, this PPAR, by way of alternative splicing, is expressed in three forms: ** γ1 - expressed in virtually all tissues, including heart, muscle, colon, kidney, pancreas, and spleen ** γ2 - expressed mainly in adipose tissue; it is 30 amino acids longer than γ1 ** γ3 - expressed in macrophage ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcriptional Regulation
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA ( transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology. The regulation of transcription is a vital process in all living organisms. It is orchestrated by transcription factors and other proteins working in concert to finely tune t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
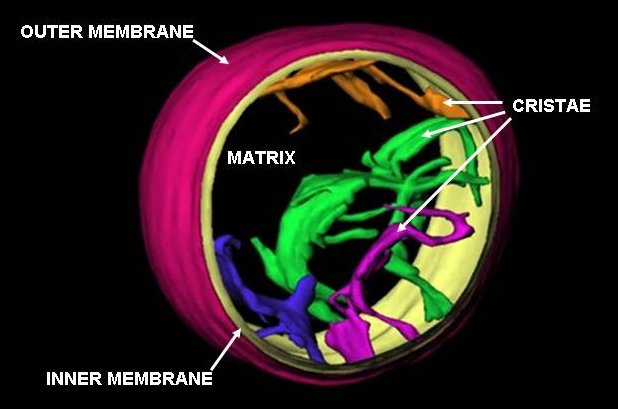
Mitochondrion
A mitochondrion () is an organelle found in the cell (biology), cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double lipid bilayer, membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'', meaning a thread-like granule, was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase popularized by Philip Siekevitz in a 1957 ''Scientific American'' article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). The multicellular animal ''Henneguya zschokkei, Henneguya salminicola'' is known to have retained mitochondrion-related organelles despite a complete loss of their mitochondrial genome. A large number ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endoplasmic Reticulum
The endoplasmic reticulum (ER) is a part of a transportation system of the eukaryote, eukaryotic cell, and has many other important functions such as protein folding. The word endoplasmic means "within the cytoplasm", and reticulum is Latin for "little net". It is a type of organelle made up of two subunits – rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae (in the RER), and tubular structures in the SER. The membranes of the ER are continuous with the outer nuclear membrane. The endoplasmic reticulum is not found in red blood cells, or spermatozoa. There are two types of ER that share many of the same proteins and engage in certain common activities such as the synthesis of certain lipids and cholesterol. Different types of Cell (biology), cells contain different ratios of the two types of ER dependin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Trafficking
Protein targeting or protein sorting is the biological mechanism by which proteins are transported to their appropriate destinations within or outside the cell. Proteins can be targeted to the inner space of an organelle, different intracellular membranes, the plasma membrane, or to the exterior of the cell via secretion. Information contained in the protein itself directs this delivery process. Correct sorting is crucial for the cell; errors or dysfunction in sorting have been linked to multiple diseases. History In 1970, Günter Blobel conducted experiments on protein translocation across membranes. Blobel, then an assistant professor at Rockefeller University, built upon the work of his colleague George Palade. Palade had previously demonstrated that non-secreted proteins were translated by free ribosomes in the cytosol, while secreted proteins (and target proteins, in general) were translated by ribosomes bound to the endoplasmic reticulum (ER). Candidate explanations at th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thiolase
Thiolases, also known as acetyl-coenzyme A acetyltransferases (ACAT), are enzymes which convert two units of acetyl-CoA to acetoacetyl CoA in the mevalonate pathway. Thiolases are ubiquitous enzymes that have key roles in many vital biochemical pathways, including the beta oxidation pathway of fatty acid degradation and various biosynthetic pathways. Members of the thiolase family can be divided into two broad categories: degradative thiolases (EC 2.3.1.16) and biosynthetic thiolases (EC 2.3.1.9). These two different types of thiolase are found both in eukaryotes and in prokaryotes: acetoacetyl-CoA thiolase (EC:2.3.1.9) and 3-ketoacyl-CoA thiolase (EC:2.3.1.16). 3-ketoacyl-CoA thiolase (also called thiolase I) has a broad chain-length specificity for its substrates and is involved in degradative pathways such as fatty acid beta-oxidation. Acetoacetyl-CoA thiolase (also called thiolase II) is specific for the thiolysis of acetoacetyl-CoA and involved in biosynthetic pathways ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |