|
Replication Protein A
Replication protein A (RPA) is the major protein that binds to single-stranded DNA (ssDNA) in eukaryotic cells. In vitro, RPA shows a much higher affinity for ssDNA than RNA or double-stranded DNA. RPA is required in replication, recombination and repair processes such as nucleotide excision repair and homologous recombination. It also plays roles in responding to damaged DNA. Structure RPA is a heterotrimer, composed of the subunits RPA1 (RPA70) (70kDa subunit), RPA2 (RPA32) (32kDa subunit) and RPA3 (RPA14) (14kDa subunit). The three RPA subunits contain six OB-folds (oligonucleotide/oligosaccharide binding), with DNA-binding domains (DBD) designated DBDs A-F, that bind RPA to single-stranded DNA. DBDs A, B, C and F are located on RPA1, DBD D is located on RPA2, and DBD E is located on RPA3. DBDs C, D, and E make up the trimerization core of the protein with flexible linker regions connecting them all together. Due to these flexible linker regions RPA is consider ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Replication Protein A1
Replication protein A 70 kDa DNA-binding subunit is a protein that in humans is encoded by the ''RPA1'' gene. Interactions Replication protein A1 has been shown to interact with: * BRCA2, * BLM, * MCM2, * MCM4, * MCM6, * MCM7, * MUTYH, * ORC2L, * ORC6L, * P53, * RPA2, * RPA3, * TIPIN, * TP53BP1, and * XPA. See also * DNA-binding subunit * Replication protein A * Replication protein A2 * Replication protein A3 * Single-stranded binding protein Single-stranded binding proteins (SSBs) are a class of proteins that have been identified in both viruses and organisms from bacteria to humans. Viral SSB Although the overall picture of ''human cytomegalovirus'' (HHV-5) DNA synthesis appears ... References Further reading * * * * * * * * * * * * * * * * * * {{DNA replication ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
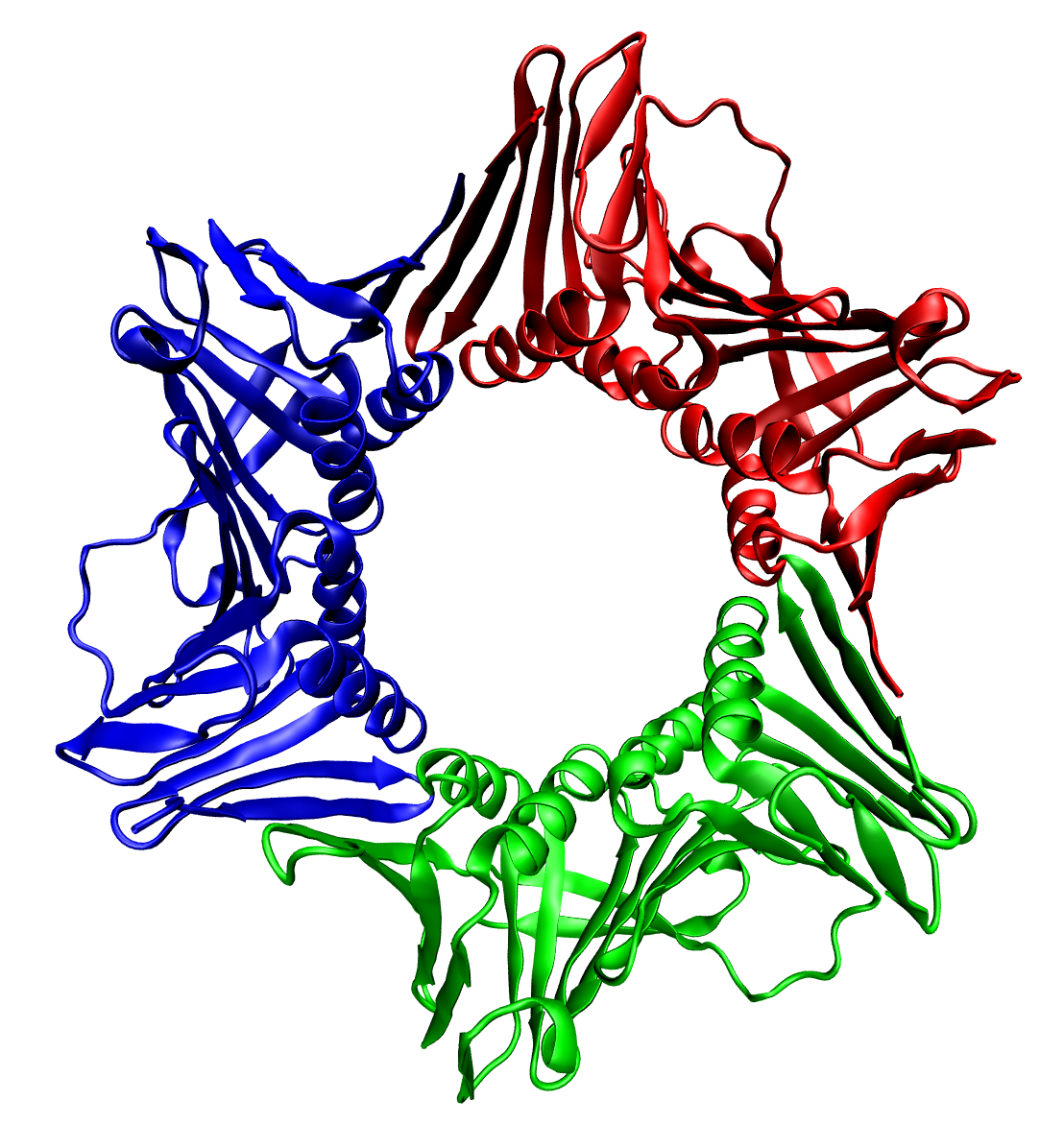
Protein Trimer
image:4tsv bio r 250.jpg, thumbnail, 400px, Trimeric form of a TNF-α mutant In biochemistry, a protein trimer is a Macromolecule, macromolecular Complex (chemistry), complex formed by three, usually covalent bond, non-covalently bound, macromolecules like proteins or nucleic acids. A protein trimer often occurs from the assembly of a protein's Protein quaternary structure, quaternary structure. The non-covalent interactions between the Hydrophobe, hydrophobic and Hydrophile, hydrophilic regions on the polypeptides units help to stabilize the quaternary structure. Since a protein trimer is composed of multiple polypeptide subunits, it is considered an oligomer. A homotrimer would be formed by three identical molecules. A heterotrimer would be formed by three different macromolecules. Type II Collagen is an example of homotrimeric protein, while Type I collagen is an AAB-type heterotrimeric protein. An example of viral protein homotrimeric protein is mammarenavirus of Z matrix prote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Meiosis
Meiosis () is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, the sperm or egg cells. It involves two rounds of division that ultimately result in four cells, each with only one copy of each chromosome (haploid). Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and a female will fuse to create a zygote, a cell with two copies of each chromosome. Errors in meiosis resulting in aneuploidy (an abnormal number of chromosomes) are the leading known cause of miscarriage and the most frequent genetic cause of developmental disabilities. In meiosis, DNA replication is followed by two rounds of cell division to produce four daughter cells, each with half the number of chromosomes as the original parent cell. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in Cell (biology), cells, by internal metabolism, metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for Transcription (biology), transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase
In biochemistry, a polymerase is an enzyme (Enzyme Commission number, EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using Base pair, base-pairing interactions or RNA by half ladder replication. A DNA polymerase from the thermophile, thermophilic bacterium, ''Thermus aquaticus'' (''Taq'') (Protein Data Bank, PDB]1BGX EC 2.7.7.7) is used in the polymerase chain reaction, an important technique of molecular biology. A polymerase may be template-dependent or template-independent. Polynucleotide adenylyltransferase, Poly-A-polymerase is an example of template independent polymerase. Terminal deoxynucleotidyl transferase also known to have template independent and template dependent activities. By function *DNA polymerase (DNA-directed DNA polymerase, DdDP) **Family A: DNA polymerase I; Pol POLG, γ, POLQ, θ, DNA polymer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endonuclease
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain (namely DNA or RNA). Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (with regard to sequence), while many, typically called '' restriction endonucleases'' or ''restriction enzymes'', cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (''endo'') portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity. Restriction enzymes are endonucleases from eubacteria and archaea that recognize a specific DNA sequence. The nucleotide sequence recognized for cleavage by a restriction enzyme is called the ''restriction site''. Typically, a restriction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stoichiometry
Stoichiometry () is the relationships between the masses of reactants and Product (chemistry), products before, during, and following chemical reactions. Stoichiometry is based on the law of conservation of mass; the total mass of reactants must equal the total mass of products, so the relationship between reactants and products must form a ratio of positive integers. This means that if the amounts of the separate reactants are known, then the amount of the product can be calculated. Conversely, if one reactant has a known quantity and the quantity of the products can be empirically determined, then the amount of the other reactants can also be calculated. This is illustrated in the image here, where the unbalanced equation is: : : However, the current equation is imbalanced. The reactants have 4 hydrogen and 2 oxygen atoms, while the product has 2 hydrogen and 3 oxygen. To balance the hydrogen, a coefficient of 2 is added to the product H2O, and to fix the imbalance of oxygen, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CST Complex
The CST complex is a cellular multiprotein complex involved in telomere maintenance. In budding yeast (''Saccharomyces cerevisiae''), it is composed of the proteins Cdc13, Stn1, and Ten1; in mammals, it consists of the proteins CTC1, STN1, and TEN1. It is related to the replication protein A complex. Structure For budding yeast as well as for mammals, CST is a protein heterotrimer, consisting of three distinct proteins. Yeast Stn1 and Ten1 are orthologous proteins to mammalian STN1 and TEN1. But yeast Cdc13 and mammalian CTC1 are very different in amino acid sequence, length, and to some extent in function. Function For both budding yeast and mammals, the CST complex contributes to telomere maintenance, but this function is more crucial for budding yeast, where the CST complex performs the functions that shelterin performs in vertebrates. At least four factors contribute to telomere maintenance: telomerase, shelterin, TERRA and the CST Complex. CST protection of telomeres for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols: : This equation can be written in several ways that are nearly equivalent that describe the behaviors of various protonated states of ATP, ADP, and the phosphorylated product. As is clear from the equation, a phosphate group per se is not transferred, but a phosphoryl group (PO3-). Phosphoryl is an electrophile. This process and its inverse, dephosphorylation, are common in biology. Text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. Protein phosphorylation often activates (or deactivates) many enzymes. During respiration Phosphorylation is essential to the processes of both anaerobic and aerobic respiration, which involve the production of adenosine triphosphate (ATP), the "high-energy" exc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ... or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Flexible Linker
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like aggregates, or flexible linkers in large multi- domain proteins. They are sometimes considered as a separate class of proteins along with globular, fibrous and membrane proteins. IDPs are a very large and functionally important class of proteins. They are most numerous in eukaryotes, with an estimated 30-40% of residues in the eukaryotic proteome located in disordered regions. Disorder is present in around 70% of proteins, either in the form of disordered tails or flexible linkers. Proteins can also be entirely disordered and lack a defined secondary and/or tertiary structure. Their discovery has disproved the idea that three-dimensional struct ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |