|
Reductive Evolution
Reductive evolution is the process by which microorganisms remove genes from their genome. It can occur when bacteria found in a free-living state enter a restrictive state (either as endosymbionts or parasites) or are completely absorbed by another organism becoming intracellular ( symbiogenesis). The bacteria will adapt to survive and thrive in the restrictive state by altering and reducing its genome to get rid of the newly redundant pathways that are provided by the host. In an endosymbiont or symbiogenesis relationship where both the guest and host benefit, the host can also undergo reductive evolution to eliminate pathways that are more efficiently provided for by the guest. Examples Endosymbiont or parasitic microorganisms such as '' Rickettsia prowazekii'', '' Chlorella'' in '' Paramecium'', '' Buchnera aphidicola'' in aphids, and '' Wolbachia'' bacteria in '' Wuchereria bancrofti'' have all been studied and fully sequenced which is why they are used as examples of r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microorganism
A microorganism, or microbe,, ''mikros'', "small") and ''organism'' from the el, ὀργανισμός, ''organismós'', "organism"). It is usually written as a single word but is sometimes hyphenated (''micro-organism''), especially in older texts. The informal synonym ''microbe'' () comes from μικρός, mikrós, "small" and βίος, bíos, "life". is an organism of microscopic size, which may exist in its single-celled form or as a colony of cells. The possible existence of unseen microbial life was suspected from ancient times, such as in Jain scriptures from sixth century BC India. The scientific study of microorganisms began with their observation under the microscope in the 1670s by Anton van Leeuwenhoek. In the 1850s, Louis Pasteur found that microorganisms caused food spoilage, debunking the theory of spontaneous generation. In the 1880s, Robert Koch discovered that microorganisms caused the diseases tuberculosis, cholera, diphtheria, and anthrax. Bec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ecological Niche
In ecology, a niche is the match of a species to a specific environmental condition. Three variants of ecological niche are described by It describes how an organism or population responds to the distribution of resources and competitors (for example, by growing when resources are abundant, and when predators, parasites and pathogens are scarce) and how it in turn alters those same factors (for example, limiting access to resources by other organisms, acting as a food source for predators and a consumer of prey). "The type and number of variables comprising the dimensions of an environmental niche vary from one species to another ndthe relative importance of particular environmental variables for a species may vary according to the geographic and biotic contexts". See also Chapter 2: Concepts of niches, pp. 7 ''ff'' A Grinnellian niche is determined by the habitat in which a species lives and its accompanying behavioral adaptations. An Eltonian niche emphasizes that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C Value Enigma
C-value is the amount, in picograms, of DNA contained within a haploid nucleus (e.g. a gamete) or one half the amount in a diploid somatic cell of a eukaryotic organism. In some cases (notably among diploid organisms), the terms C-value and genome size are used interchangeably; however, in polyploids the C-value may represent two or more genomes contained within the same nucleus. Greilhuber ''et al.'' have suggested some new layers of terminology and associated abbreviations to clarify this issue, but these somewhat complex additions are yet to be used by other authors. Origin of the term Many authors have incorrectly assumed that the 'C' in "C-value" refers to "characteristic", "content", or "complement". Even among authors who have attempted to trace the origin of the term, there had been some confusion because Hewson Swift did not define it explicitly when he coined it in 1950. In his original paper, Swift appeared to use the designation "1C value", "2C value", etc., in r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Size
Genome size is the total amount of DNA contained within one copy of a single complete genome. It is typically measured in terms of mass in picograms (trillionths (10−12) of a gram, abbreviated pg) or less frequently in daltons, or as the total number of nucleotide base pairs, usually in megabases (millions of base pairs, abbreviated Mb or Mbp). One picogram is equal to 978 megabases. In diploid organisms, genome size is often used interchangeably with the term C-value. An organism's complexity is not directly proportional to its genome size; total DNA content is widely variable between biological taxa. Some single-celled organisms have much more DNA than humans, for reasons that remain unclear (see non-coding DNA and C-value enigma). Origin of the term The term "genome size" is often erroneously attributed to a 1976 paper by Ralph Hinegardner, even in discussions dealing specifically with terminology in this area of research (e.g., Greilhuber 2005). Notably, Hinegar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Maximum Likelihood Sequence Estimation
Maximum likelihood sequence estimation (MLSE) is a mathematical algorithm to extract useful data out of a noisy data stream. Theory For an optimized detector for digital signals the priority is not to reconstruct the transmitter signal, but it should do a best estimation of the transmitted data with the least possible number of errors. The receiver emulates the distorted channel. All possible transmitted data streams are fed into this distorted channel model. The receiver compares the time response with the actual received signal and determines the most likely signal. In cases that are most computationally straightforward, root mean square deviation can be used as the decision criterionG. Bosco, P. Poggiolini, and M. Visintin, "Performance Analysis of MLSE Receivers Based on the Square-Root Metric," J. Lightwave Technol. 26, 2098–2109 (2008) for the lowest error probability. Background Suppose that there is an underlying signal , of which an observed signal is available. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Maximum Parsimony (phylogenetics)
In phylogenetics, maximum parsimony is an optimality criterion under which the phylogenetic tree that minimizes the total number of character-state changes (or miminizes the cost of differentially weighted character-state changes) is preferred. Under the maximum-parsimony criterion, the optimal tree will minimize the amount of homoplasy (i.e., convergent evolution, parallel evolution, and evolutionary reversals). In other words, under this criterion, the shortest possible tree that explains the data is considered best. Some of the basic ideas behind maximum parsimony were presented by James S. Farris in 1970 and Walter M. Fitch in 1971. Maximum parsimony is an intuitive and simple criterion, and it is popular for this reason. However, although it is easy to ''score'' a phylogenetic tree (by counting the number of character-state changes), there is no algorithm to quickly ''generate'' the most-parsimonious tree. Instead, the most-parsimonious tree must be sought in "tree s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lynn Margulis
Lynn Margulis (born Lynn Petra Alexander; March 5, 1938 – November 22, 2011) was an American evolutionary biologist, and was the primary modern proponent for the significance of symbiosis in evolution. Historian Jan Sapp has said that "Lynn Margulis's name is as synonymous with symbiosis as Charles Darwin's is with evolution." In particular, Margulis transformed and fundamentally framed current understanding of the evolution of cells with nuclei – an event Ernst Mayr called "perhaps the most important and dramatic event in the history of life" – by proposing it to have been the result of symbiotic mergers of bacteria. Margulis was also the co-developer of the Gaia hypothesis with the British chemist James Lovelock, proposing that the Earth functions as a single self-regulating system, and was the principal defender and promulgator of the five kingdom classification of Robert Whittaker. Throughout her career, Margulis' work could arouse intense objection (one grant appli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Sizes
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of 'junk' DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been sequenced and various regions have been annotated. The International Human Genome Project reported the sequence of the genome for ''Homo sapiens'' in 200The Human Genome Project although the initial "finished" sequence was missing 8% of the genome consisting mostly of repetitive sequences. With advancements in technology that could handle sequencin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
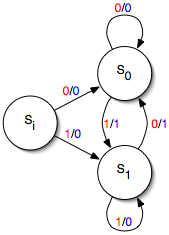
Mealy Bug
In the theory of computation, a Mealy machine is a finite-state machine whose output values are determined both by its current state and the current inputs. This is in contrast to a Moore machine, whose output values are determined solely by its current state. A Mealy machine is a deterministic finite-state transducer: for each state and input, at most one transition is possible. History The Mealy machine is named after George H. Mealy, who presented the concept in a 1955 paper, "A Method for Synthesizing Sequential Circuits". Formal definition A Mealy machine is a 6-tuple (S, S_0, \Sigma, \Lambda, T, G) consisting of the following: * a finite set of states S * a start state (also called initial state) S_0 which is an element of S * a finite set called the input alphabet \Sigma * a finite set called the output alphabet \Lambda * a transition function T : S \times \Sigma \rightarrow S mapping pairs of a state and an input symbol to the corresponding next state. * an output functi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Candidatus Tremblaya
In prokaryote nomenclature, ''Candidatus'' (Latin for candidate of Roman office) is used to name prokaryotic phyla that are well characterized but yet-uncultured. Contemporary sequencing approaches, such as 16S sequencing or metagenomics, provide much information about the analyzed organisms and thus allow to identify and characterize individual species. However, the majority of prokaryotic species remain uncultivable and hence inaccessible for further characterization in ''in vitro'' study. The recent discoveries of a multitude of candidate taxa has led to candidate phyla radiation expanding the tree of life through the new insights in bacterial diversity. Nomenclature History The initial International Code of Nomenclature of Prokaryotes as well as early revisions did not account for the possibility of identifying prokaryotes which were not yet cultivable. Therefore, the term ''Candidatus'' was proposed in the context of a conference of the International Committee on Systemati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guanine
Guanine () (symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine ( uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is called guanosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine- imidazole ring system with conjugated double bonds. This unsaturated arrangement means the bicyclic molecule is planar. Properties Guanine, along with adenine and cytosine, is present in both DNA and RNA, whereas thymine is usually seen only in DNA, and uracil only in RNA. Guanine has two tautomeric forms, the major keto form (see figures) and rare enol form. It binds to cytosine through three hydrogen bonds. In cytosine, the amino group acts as the hydrogen bond donor and the C-2 carbonyl and the N-3 amine as the hydrogen-bond acceptors. Guanine has the C-6 carbonyl group that acts as the hydrogen bond acceptor, while a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |