|
RaiA RNA Motif
The ''raiA'' RNA motif is a conserved RNA structure that was discovered by bioinformatics. ''raiA'' motif RNAs are found in Actinomycetota and Bacillota, and have many conserved features—including conserved nucleotide positions, conserved secondary structures and associated protein-coding genes—in both of these phyla. Some conserved features of the ''raiA'' RNA motif suggest that they function as cis-regulatory elements, but other aspects of the motif suggest otherwise. ''raiA'' RNAs usually occur upstream of protein-coding genes. Although this association could suggest a ''cis''-regulatory function, the ''raiA'' RNAs are fairly often located unusually far (more than 600 bp) from the genes compared to typical ''cis''-regulatory RNAs. Moreover, although most ''raiA'' RNAs are located upstream of protein-coding genes, many are not. In both Bacillota and Actinomycetota, ''raiA'' RNAs commonly occur upstream of genes encoding RaiA, a protein that binds to the ribosome as par ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes. During gene expression (the synthesis of Gene product, RNA or protein from a gene), DNA is first transcription (biology), copied into RNA. RNA can be non-coding RNA, directly functional or be the intermediate protein biosynthesis, template for the synthesis of a protein. The transmission of genes to an organism's offspring, is the basis of the inheritance of phenotypic traits from one generation to the next. These genes make up different DNA sequences, together called a genotype, that is specific to every given individual, within the gene pool of the population (biology), population of a given species. The genotype, along with environmental and developmental factors, ultimately determines the phenotype ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Untranslated Region
In molecular genetics, an untranslated region (or UTR) refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the Directionality (molecular biology), 5' side, it is called the Five prime untranslated region, 5' UTR (or leader sequence), or if it is found on the Directionality (molecular biology), 3' side, it is called the Three prime untranslated region, 3' UTR (or trailer sequence). mRNA is RNA that carries information from DNA to the ribosome, the site of protein synthesis (Translation (biology), translation) within a cell. The mRNA is initially Transcription (genetics), transcribed from the corresponding DNA sequence and then translated into protein. However, several regions of the mRNA are usually not translated into protein, including the 5' and 3' UTRs. Although they are called untranslated regions, and do not form the Coding region, protein-coding region of the gene, Upstream open reading frame, uORFs located withi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Natural Competence
In microbiology, genetics, cell biology, and molecular biology, competence is the ability of a cell to alter its genetics by taking up extracellular DNA from its environment through a process called transformation. Competence can be differentiated between natural competence and induced or artificial competence. Natural competence is a genetically specified ability of bacteria that occurs under natural conditions as well as in the laboratory. Artificial competence arises when cells in laboratory cultures are treated to make them transiently permeable to DNA. Competence allows for rapid adaptation and DNA repair of the cell. History Natural competence was discovered by Frederick Griffith in 1928, when he showed that a preparation of killed cells of a pathogenic bacterium contained something that could transform related non-pathogenic cells into the pathogenic type. In 1944 Oswald Avery, Colin MacLeod, and Maclyn McCarty demonstrated that this 'transforming factor' was pure DNA. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
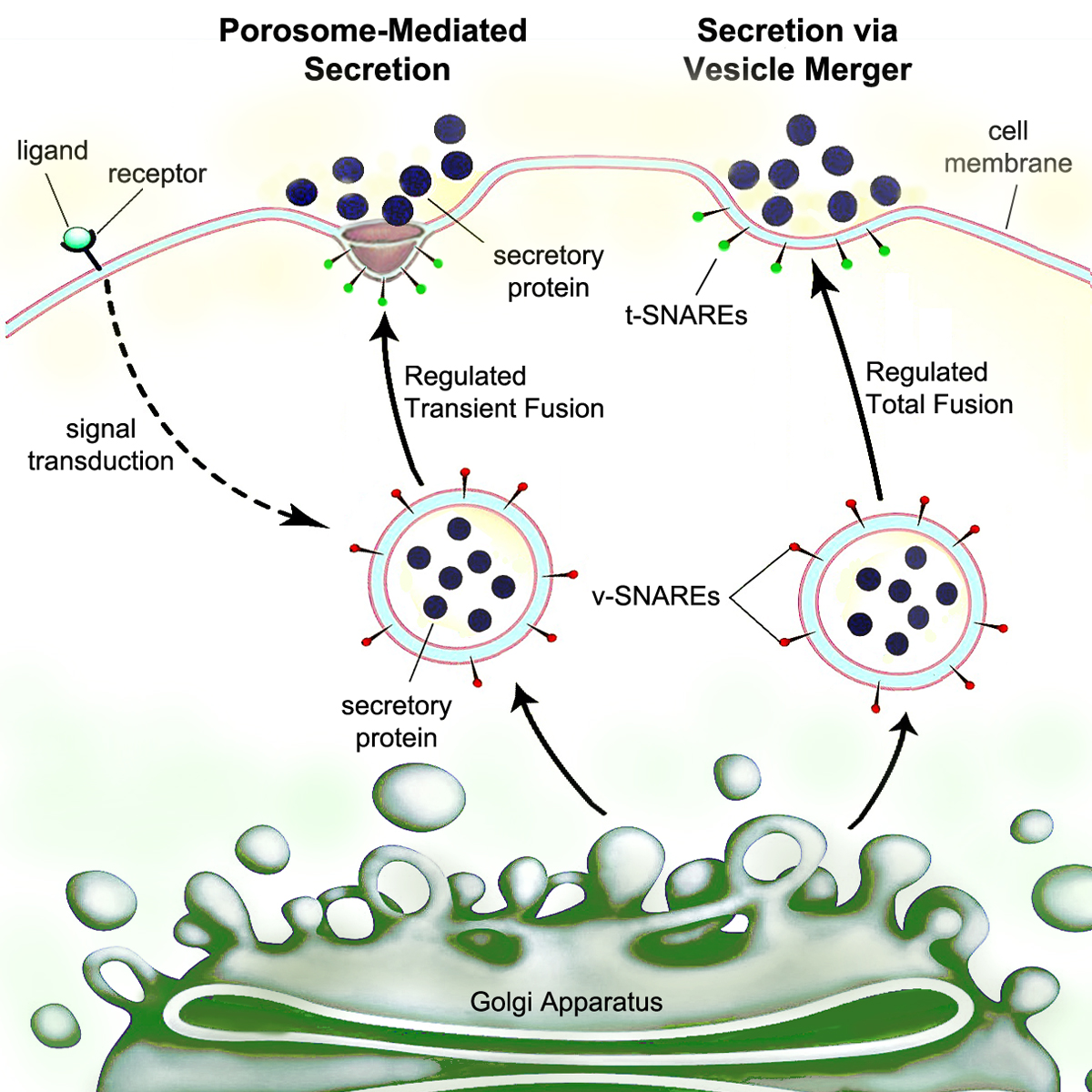
Secretion
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structures embedded in the cell membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell. Secretion in bacterial species means the transport or translocation of effector molecules. For example: proteins, enzymes or toxins (such as cholera toxin in pathogenic bacteria e.g. '' Vibrio cholerae'') from across the interior (cytoplasm or cytosol) of a bacterial cell to its exterior. Secretion is a very important mechanism in bacterial functioning and operation in their natural surrounding environment for adaptation and survival. In eukaryotic cells ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA molecules and many ribosomal proteins (). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA (mRNA) chain. Ribosomes bind to the messenger RNA molecules and use the RNA's sequence of nucleotides to determine the sequence of amino acids needed to generate a protein. Amino acids are selected and carried to the ribosome by transfer RNA (tRNA) molecules, which enter the riboso ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Elements
''Cis''-regulatory elements (CREs) or ''cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes (pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are typi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylum
In biology, a phylum (; : phyla) is a level of classification, or taxonomic rank, that is below Kingdom (biology), kingdom and above Class (biology), class. Traditionally, in botany the term division (taxonomy), division has been used instead of phylum, although the International Code of Nomenclature for algae, fungi, and plants accepts the terms as equivalent. Depending on definitions, the animal kingdom Animalia contains about 31 phyla, the plant kingdom Plantae contains about 14 phyla, and the fungus kingdom Fungi contains about eight phyla. Current research in phylogenetics is uncovering the relationships among phyla within larger clades like Ecdysozoa and Embryophyta. General description The term phylum was coined in 1866 by Ernst Haeckel from the Greek (, "race, stock"), related to (, "tribe, clan"). Haeckel noted that species constantly evolved into new species that seemed to retain few consistent features among themselves and therefore few features that distinguishe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metabolic reactions, DNA replication, Cell signaling, responding to stimuli, providing Cytoskeleton, structure to cells and Fibrous protein, organisms, and Intracellular transport, transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the Nucleic acid sequence, nucleotide sequence of their genes, and which usually results in protein folding into a specific Protein structure, 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called pep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids (DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst eukaryotes, and the tmRNA in bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, promp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Secondary Structure
Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as a list of bases which are paired in a nucleic acid molecule. The secondary structures of biological DNAs and RNAs tend to be different: biological DNA mostly exists as fully base paired double helices, while biological RNA is single stranded and often forms complex and intricate base-pairing interactions due to its increased ability to form hydrogen bonds stemming from the extra hydroxyl group in the ribose sugar. In a non-biological context, secondary structure is a vital consideration in the nucleic acid design of nucleic acid structures for DNA nanotechnology and DNA computing, since the pattern of basepairing ultimately determines the overall structure of the molecules. Fundamental concepts Base pairing In molecular biology, two nucleotides on opposite complementary DNA or RNA strands that are connected via hydroge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |