|
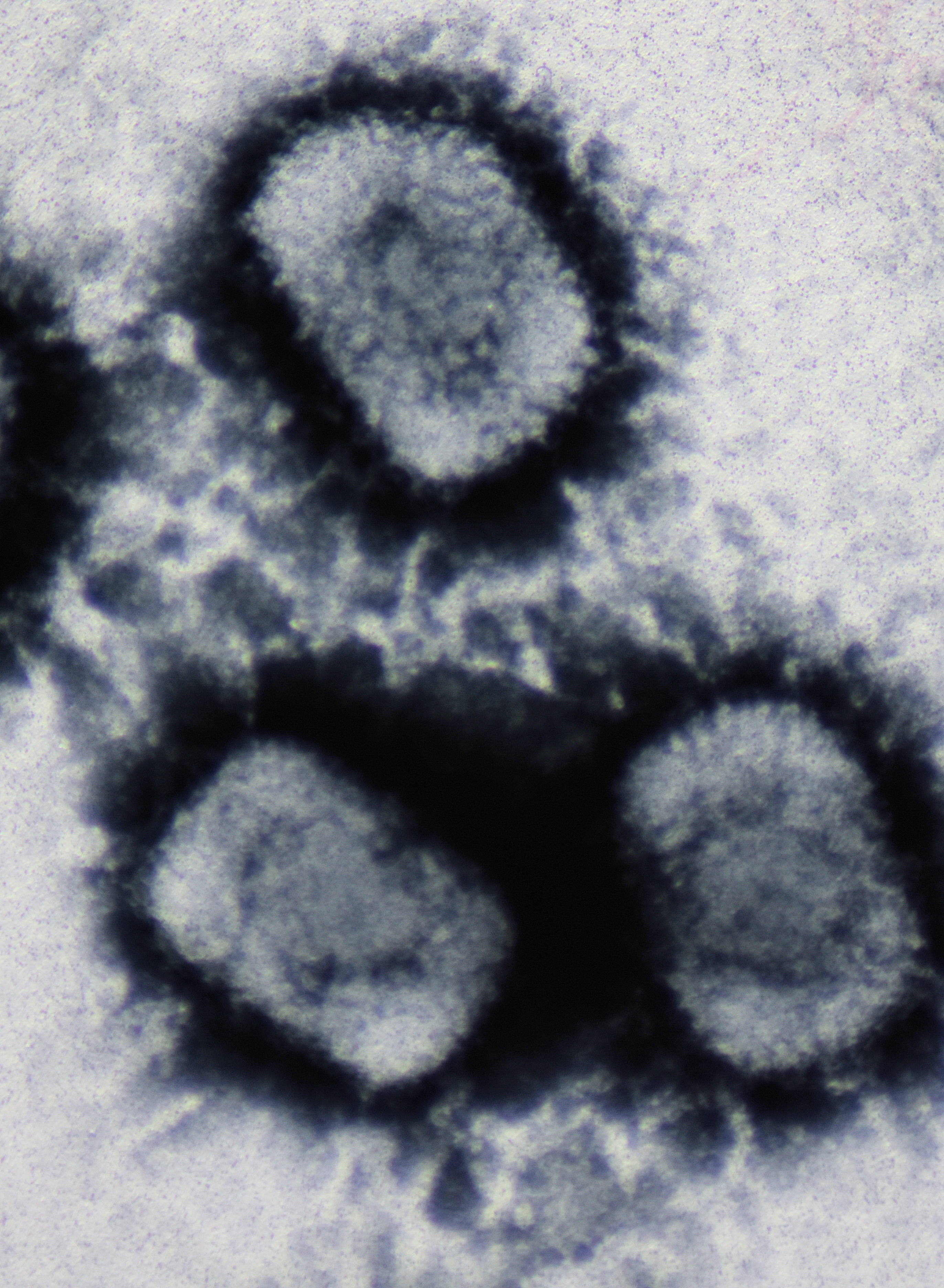
Raccoonpox Virus
Raccoonpox virus (RCN) is a double-stranded DNA virus and a member of the ''Orthopoxvirus'' genus in the family ''Poxviridae'' and subfamily ''Chordopoxvirinae.'' Vertebrates are the natural host of Chordopoxvirinae subfamily viruses.Pastoret, P. P., & Vanderplasschen, A. (2003). Poxviruses as vaccine vectors lectronic version Comparative Immunology, Microbiology, and Infectious Diseases, 26, 343-355. More specifically, raccoons are the natural hosts of RCN. RCN was isolated in 1961 from the upper respiratory tissues of 2 raccoons in a group of 92 observably healthy raccoons (Procyon lotor) trapped close to Aberdeen, Maryland. Of the 92 apparently healthy raccoons trapped near Aberdeen, Maryland in 1961, 22 had sera containing RCN HA1 antibodies. The sera partially cross-reacted with a vaccinia virus HA preparation, suggesting a close relation between the viruses. Unlike the HA of other vaccinia-like viruses, the HA of RCN did not cross-react with monkeypox virus HA. Though RP ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Virus
A DNA virus is a virus that has a genome made of deoxyribonucleic acid (DNA) that is replicated by a DNA polymerase. They can be divided between those that have two strands of DNA in their genome, called double-stranded DNA (dsDNA) viruses, and those that have one strand of DNA in their genome, called single-stranded DNA (ssDNA) viruses. dsDNA viruses primarily belong to two realms: ''Duplodnaviria'' and ''Varidnaviria'', and ssDNA viruses are almost exclusively assigned to the realm ''Monodnaviria'', which also includes some dsDNA viruses. Additionally, many DNA viruses are unassigned to higher taxa. Reverse transcribing viruses, which have a DNA genome that is replicated through an RNA intermediate by a reverse transcriptase, are classified into the kingdom '' Pararnavirae'' in the realm ''Riboviria''. DNA viruses are ubiquitous worldwide, especially in marine environments where they form an important part of marine ecosystems, and infect both prokaryotes and eukaryotes. They a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, reading frames are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an open reading frame (ORF) may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycosaminoglycan
Glycosaminoglycans (GAGs) or mucopolysaccharides are long, linear polysaccharides consisting of repeating disaccharide units (i.e. two-sugar units). The repeating two-sugar unit consists of a uronic sugar and an amino sugar, except in the case of the sulfated glycosaminoglycan keratan, where, in place of the uronic sugar there is a galactose unit. GAGs are found in vertebrates, invertebrates and bacteria. Because GAGs are highly polar molecules and attract water; the body uses them as lubricants or shock absorbers. Mucopolysaccharidoses are a group of metabolic disorders in which abnormal accumulations of glycosaminoglycans occur due to enzyme deficiencies. Production Glycosaminoglycans vary greatly in molecular mass, disaccharide structure, and sulfation. This is because GAG synthesis is not template driven, as are proteins or nucleic acids, but constantly altered by processing enzymes. GAGs are classified into four groups, based on their core disaccharide structures: # H ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hemagglutinin
The term hemagglutinin (alternatively spelt ''haemagglutinin'', from the Greek , 'blood' + Latin , 'glue') refers to any protein that can cause red blood cells (erythrocytes) to clump together (" agglutinate") ''in vitro''. They do this by binding to the sugar residues on a red blood cell; when a single hemagglutinin molecule binds sugars from multiple red blood cells, it "glues" these cells together. As a result, they are carbohydrate-binding proteins (lectins). The ability to bind red blood cell sugars have independently appeared several times, and as a result hemaglutinins do not all bind using the same mechanism. The ability to bind red blood sugars is also not necessarily related to the ''in vivo'' function of the protein. The term ''hemagglutinin'' is most commonly applied to plant and viral lectins. Natural proteins that clump together red blood cells were known since the turn of the 19th century. Virologist George K. Hirst is also credited for "discovering agglutination a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Fragment
In molecular biology, a restriction fragment is a DNA fragment resulting from the cutting of a DNA strand by a restriction enzyme (restriction endonucleases). Each restriction enzyme is highly specific, recognising a particular short DNA sequence (a restriction site) and cutting both DNA strands at specific points within this site. Most restriction sites are palindromic (i.e. the sequence of nucleotides is the same on both strands when read in the 5' to 3' direction of each strand) and are four to eight nucleotides long. Many cuts are made by one restriction enzyme because of the chance repetition of these sequences in a long DNA molecule, yielding a set of restriction fragments. A particular DNA molecule will always yield the same set of restriction fragments when exposed to the same restriction enzyme. Restriction fragments can be analyzed using techniques such as gel electrophoresis or used in recombinant DNA technology. Applications In recombinant DNA technology, spec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tandem Repeat
In genetics, tandem repeats occur in DNA when a pattern of one or more nucleotides is repeated and the repetitions are directly adjacent to each other, e.g. ATTCG ATTCG ATTCG, in which the sequence ATTCG is repeated three times. Several protein domains also form tandem repeats within their amino acid primary structure, such as armadillo repeats. However, in proteins, perfect tandem repeats are rare in naturally proteins, but they have been added to designed proteins. Tandem repeats constitute about 8% of the human genome. They are implicated in more than 50 lethal human diseases, including amyotrophic lateral sclerosis, Huntington's disease, and several cancers. Terminology All tandem repeat arrays are classifiable as satellite DNA, a name originating from the fact that tandem DNA repeats, by nature of repeating the same nucleotide sequences repeatedly, have a unique ratio of the two possible nucleotide base pair combinations, conferring them a specific mass density that a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymorphism (biology)
In biology, polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative '' phenotypes'', in the population of a species. To be classified as such, morphs must occupy the same habitat at the same time and belong to a panmictic population (one with random mating). Ford E.B. 1965. ''Genetic polymorphism''. Faber & Faber, London. Put simply, polymorphism is when there are two or more possibilities of a trait on a gene. For example, there is more than one possible trait in terms of a jaguar's skin colouring; they can be light morph or dark morph. Due to having more than one possible variation for this gene, it is termed 'polymorphism'. However, if the jaguar has only one possible trait for that gene, it would be termed "monomorphic". For example, if there was only one possible skin colour that a jaguar could have, it would be termed monomorphic. The term polyphenism can be used to clarify that the different forms arise from the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA enzyme substrate (biology), substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each backbone chain, sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GC-content
In molecular biology and genetics, GC-content (or guanine-cytosine content) is the percentage of nitrogenous bases in a DNA or RNA molecule that are either guanine (G) or cytosine (C). This measure indicates the proportion of G and C bases out of an implied four total bases, also including adenine and thymine in DNA and adenine and uracil in RNA. GC-content may be given for a certain fragment of DNA or RNA or for an entire genome. When it refers to a fragment, it may denote the GC-content of an individual gene or section of a gene (domain), a group of genes or gene clusters, a non-coding region, or a synthetic oligonucleotide such as a primer. Structure Qualitatively, guanine (G) and cytosine (C) undergo a specific hydrogen bonding with each other, whereas adenine (A) bonds specifically with thymine (T) in DNA and with uracil (U) in RNA. Quantitatively, each GC base pair is held together by three hydrogen bonds, while AT and AU base pairs are held together by two hydrogen bonds ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loops are nucleic acid Biomolecular structure, secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regions of the same nucleic acid strand, usually Complementarity (molecular biology), complementary in nucleotide sequence, base-pair to form a double helix that ends in a loop of unpaired nucleotides. Stem-loops are most commonly found in RNA, and are a key building block of many RNA biomolecular structure#Secondary structure, secondary structures. Stem-loops can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA-binding protein, RNA binding proteins, and serve as a Substrate (chemistry), substrate for Enzyme catalysis, enzymatic reactions. Formation and stability The formation of a stem-loop is dependent on the stability of the helix and loop regions. The first prerequisite is the p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monopartite
Monopartite refers to the class of genome that is presented in the genome of the virus. As opposed to multipartite, viruses composed of monopartite genomes have a single molecule of nucleic acid. Most dsDNA Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ... viruses are monopartite. References Genomics {{Virus-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virion
A virion (plural, ''viria'' or ''virions'') is an inert virus particle capable of invading a Cell (biology), cell. Upon entering the cell, the virion disassembles and the genetic material from the virus takes control of the cell infrastructure, thus enabling the virus to Replication (virus), replicate. The genetic material (''Viral core, core'', either DNA or RNA, along with occasionally present virus core protein) inside the virion is usually enclosed in a protection shell, known as the capsid. While the terms "virus" and "virion" are occasionally confused, recently "virion" is used solely to describe the virus structure outside of cells, while the terms "virus/viral" are broader and also include biological properties such as the infectivity of a virion. Components A virion consists of one or more nucleic acid genome molecules (single-stranded or double-stranded RNA or DNA) and coatings (a capsid and possibly a viral envelope). The virion may contain other proteins (for examp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |