|
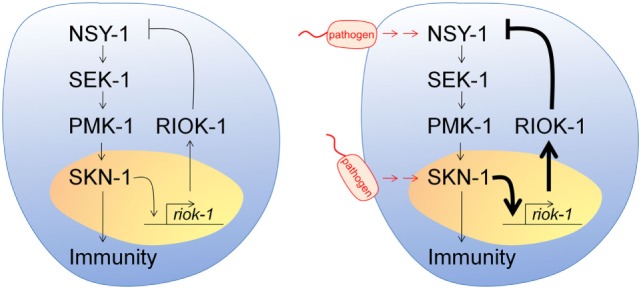
RIOK1
Serine/threonine-protein kinase RIO1 is an enzyme that in humans is encode by the RIOK1 gene. RIOK1 is an atypical protein, which exists in most archaea and eukaryotes. It belongs to the serine/threonine-specific protein kinase family. It has been intensely studied to understand the maturation they promote on small ribosomal subunits (SSU). It is suggested that over-expression or mutations of the RIOK 1 gene may cause mis-regulation of its network (in metazoans - large signaling network at the protein and gene levels via which it stimulates or restricts growth and division in response to nutrient availability). This was observed in primary cancer cells and may contribute to cancer initiation and progression. Characteristics RIOK 1 has a molecular weight of 65,583 Da, a basal isoelectric point of 5.84 (predict pI for various phosphorylation states; pI for unphosphorylated state = 5.84), and a chromosomal location of human orthodox 6p24.3.6:7,389,496-7,418,037 PTM Effects Eff ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Arginine Methyltransferase 5
Protein arginine N-methyltransferase 5 is an enzyme that in humans is encoded by the ''PRMT5'' gene. PRMT5 symmetrically dimethylates H2AR3, H4R3, H3R2, and H3R8 in vivo, all of which are linked to a range of transcriptional regulatory events. PRMT5 is a highly conserved arginine methyltransferase that translocated from the cytoplasm to the nucleus at embryonic day ~E8.5, and during preimplantation development at the ~4-cell stage. Model organisms Model organisms have been used in the study of PRMT5 function. A conditional knockout mouse line, called ''Prmt5tm2a(EUCOMM)Wtsi'' was generated as part of the International Knockout Mouse Consortium program — a high-throughput mutagenesis project to generate and distribute animal models of disease to interested scientists. Male and female animals underwent a standardized phenotypic screen to determine the effects of deletion. Twenty five tests were carried out on mutant mice and two significant abnormalities were observed. No homoz ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are Ribozyme, catalytic RNA molecules, called ribozymes. Enzymes' Chemical specificity, specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a Prokaryote, prokaryotic DNA sequence, where only one of the #Six-frame translation, six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by Transcription (biology), transcription of the DNA and its subsequent interaction with the ribosome in Translation (biology), translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription terminator, transcription termination site is located after the ORF, beyond the Translation (biology), translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, genet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Primary Structure
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences. Formation Biological Amino acids are polymerised via peptide bonds to form a long backbone, with the different amino acid side chains protruding along it. In biological systems, proteins are produced during translation by a cell's ribosomes. Some organisms can also make short peptides by non-ribosomal peptide synthesis, which often use amino acids other than the standard 20, and may be cyclised, modified and cross-linked. Chemical Peptides can be synthesised chemically via a range of laboratory methods. Chemical methods typically syn ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. The main components of the cytoplasm are cytosol (a gel-like substance), the organelles (the cell's internal sub-structures), and various cytoplasmic inclusions. The cytoplasm is about 80% water and is usually colorless. The submicroscopic ground cell substance or cytoplasmic matrix which remains after exclusion of the cell organelles and particles is groundplasm. It is the hyaloplasm of light microscopy, a highly complex, polyphasic system in which all resolvable cytoplasmic elements are suspended, including the larger organelles such as the ribosomes, mitochondria, the plant plastids, lipid droplets, and vacuoles. Most cellular activities take place within the cytoplasm, such as many metabolic pathways including glycolysis, and proc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. Protein phosphorylation often activates (or deactivates) many enzymes. Glucose Phosphorylation of sugars is often the first stage in their catabolism. Phosphorylation allows cells to accumulate sugars because the phosphate group prevents the molecules from diffusing back across their transporter. Phosphorylation of glucose is a key reaction in sugar metabolism. The chemical equation for the conversion of D-glucose to D-glucose-6-phosphate in the first step of glycolysis is given by :D-glucose + ATP → D-glucose-6-phosphate + ADP :ΔG° = −16.7 kJ/mol (° indicates measurement at standard condition) Hepatic cells are freely permeable to glucose, an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SSU RRNA
Small subunit ribosomal ribonucleic acid (SSU rRNA) is the smaller of the two major RNA components of the ribosome. Associated with a number of ribosomal protein A ribosomal protein (r-protein or rProtein) is any of the proteins that, in conjunction with rRNA, make up the ribosomal subunits involved in the cellular process of translation. ''E. coli'', other bacteria and Archaea have a 30S small subunit ...s, the SSU rRNA forms the small subunit of the ribosome. It is encoded by SSU- rDNA. Characteristics Use in phylogenetics SSU rRNA sequences are widely used for determining evolutionary relationships among organisms, since they are of ancient origin and are found in all known forms of life. See also * LSU rRNA: the large subunit ribosomal ribonucleic acid. References {{Ribosome subunits Ribosomal RNA Protein biosynthesis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrolase
Hydrolase is a class of enzyme that commonly perform as biochemical catalysts that use water to break a chemical bond, which typically results in dividing a larger molecule into smaller molecules. Some common examples of hydrolase enzymes are esterases including lipases, phosphatases, glycosidases, peptidases, and nucleosidases. Esterases cleave ester bonds in lipids and phosphatases cleave phosphate groups off molecules. An example of crucial esterase is acetylcholine esterase, which assists in transforming the neuron impulse into the acetate group after the hydrolase breaks the acetylcholine into choline and acetic acid. Acetic acid is an important metabolite in the body and a critical intermediate for other reactions such as glycolysis. Lipases hydrolyze glycerides. Glycosidases cleave sugar molecules off carbohydrates and peptidases hydrolyze peptide bonds. Nucleosidases hydrolyze the bonds of nucleotides. Hydrolase enzymes are important for the body because they h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATP-binding Motif
An ATP-binding motif is a 250-residue sequence within an ATP-binding protein’s primary structure. The binding motif is associated with a protein’s structure and/or function. ATP is a molecule of energy, and can be a coenzyme, involved in a number of biological reactions. ATP is proficient at interacting with other molecules through a binding site. The ATP binding site is the environment in which ATP catalytically actives the enzyme and, as a result, is hydrolyzed to ADP. The binding of ATP causes a conformational change to the enzyme it is interacting with. The genetic and functional similarity of such a motif demonstrates micro-evolution: proteins have co-opted the same binding sequence from other enzymes rather than developing them independently. ATP binding sites, which may be representative of an ATP binding motif, are present in many proteins which require an input of energy (from ATP), such sites as active membrane transporters, microtubule subunits, flagellum proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein–protein Interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context. Proteins rarely act alone as their functions tend to be regulated. Many molecular processes within a cell are carried out by molecular machines that are built from numerous protein components organized by their PPIs. These physiological interactions make up the so-called interactomics of the organism, while aberrant PPIs are the basis of multiple aggregation-related diseases, such as Creutzfeldt–Jakob and Alzheimer's diseases. PPIs have been studied with many methods and from different perspectives: biochemistry, quantum chemistry, molecular dynamics, signal tr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |