|
Prophage
A prophage is a bacteriophage (often shortened to "phage") genome that is integrated into the circular bacterial chromosome or exists as an extrachromosomal plasmid within the bacterial cell (biology), cell. Integration of prophages into the bacterial host is the characteristic step of the lysogenic cycle of Temperateness (virology), temperate phages. Prophages remain viral latency, latent in the genome through multiple cell divisions until activation by an external factor, such as UV light, leading to production of new phage particles that will Lysis, lyse the cell and spread. As ubiquitous mobile genetic elements, prophages play important roles in bacterial genetics and evolution, such as in the acquisition of virulence factors. Prophage induction Upon detection of host cell damage by UV light or certain chemicals, the prophage is excised from the bacterial chromosome in a process called prophage induction. After induction, viral replication begins via the lytic cycle. In the l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysogenic Cycle
Lysogeny, or the lysogenic cycle, is one of two cycles of Virus, viral reproduction (the lytic cycle being the other). Lysogeny is characterized by integration of the bacteriophage nucleic acid into the host Bacteria, bacterium's genome or formation of a circular Replicon (genetics), replicon in the bacterial cytoplasm. In this condition the bacterium continues to live and reproduce normally, while the bacteriophage lies in a dormant state in the host cell. The genetic material of the bacteriophage, called a prophage, can be transmitted to daughter cells at each subsequent cell division, and later events (such as ultraviolet, UV radiation or the presence of certain chemicals) can release it, causing proliferation of new phages via the lytic cycle. Lysogenic cycles can also occur in eukaryotes, although the method of DNA incorporation is not fully understood. For instance, the HIV, HIV viruses can either infect humans lytically, or virus latency, lay dormant (lysogenic) as part ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lambda Phage
Lambda phage (coliphage λ, scientific name ''Lambdavirus lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Lederberg in 1950. The wild type of this virus has a Temperate (virology), temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell. The phage particle consists of a head (also known as a capsid), a tail, and tail fibers (see image of virus below). The head contains the phage's double-strand linear DNA genome. During infections, the phage particle recognizes and binds to its host, ''E. coli'', causing DNA in the head of the phage to be ejected through the tail into the cytopla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mobilome
The mobilome is the entire set of mobile genetic elements in a genome. Mobilomes are found in eukaryotes, prokaryotes, and viruses. The compositions of mobilomes differ among lineages of life, with transposable elements being the major mobile elements in eukaryotes, and plasmids and prophages being the major types in prokaryotes. Virophages contribute to the viral mobilome. Mobilome in eukaryotes Transposable elements are elements that can move about or propagate within the genome, and are the major constituents of the eukaryotic mobilome. Transposable elements can be regarded as genetic parasites because they exploit the host cell's transcription and translation mechanisms to extract and insert themselves in different parts of the genome, regardless of the phenotypic effect on the host. Eukaryotic transposable elements were first discovered in maize (''Zea mays'') in which kernels showed a dotted color pattern. Barbara McClintock described the maize Ac/Ds system in whi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophage
A bacteriophage (), also known informally as a phage (), is a virus that infects and replicates within bacteria. The term is derived . Bacteriophages are composed of proteins that Capsid, encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes (e.g. Bacteriophage MS2, MS2) and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm. Bacteriophages are among the most common and diverse entities in the biosphere. Bacteriophages are ubiquitous viruses, found wherever bacteria exist. It is estimated there are more than 1031 bacteriophages on the planet, more than every other organism on Earth, including bacteria, combined. Viruses are the most abundant biological entity in the water column of the world's oceans, and the second largest component of biomass after prokaryotes, where up to 9x108 virus, virions per millilitre have b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homologous Recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in Cell (biology), cellular organisms but may be also RNA in viruses). Homologous recombination is widely used by cells to accurately DNA repair, repair harmful DNA breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and ovum, egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to Adaptation, adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RecA
RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA in bacteria. Structural and functional homologs to RecA have been found in all kingdoms of life. RecA serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea. RecA has multiple activities, all related to DNA repair. In the bacterial SOS response, it functions as a co-protease in the autocatalytic cleavage of the LexA repressor and the λ repressor. Function Homologous recombination The RecA protein binds strongly and in long clusters to ssDNA to form a nucleoprotein filament. The protein has multiple DNA binding sites, and thus can hold a single strand and double strand together. This feature makes it possible to catalyze a DNA synapsis reaction between a DNA double helix and a complementary region of single-stranded DNA. The RecA-ssDNA filament searches for sequence similarity along the dsDNA. A disordere ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Conversion
Gene conversion is the process by which one DNA sequence replaces a homologous sequence such that the sequences become identical after the conversion. Gene conversion can be either allelic, meaning that one allele of the same gene replaces another allele, or ectopic, meaning that one Sequence homology#Paralogy, paralogous DNA sequence converts another. Allelic gene conversion Allelic gene conversion occurs during meiosis when homologous recombination between heterozygotic sites results in a mismatch in base pairing. This mismatch is then recognized and corrected by the cellular machinery causing one of the alleles to be converted to the other. This can cause non-Mendelian segregation of alleles in germ cells. Nonallelic/ectopic gene conversion Recombination occurs not only during meiosis, but also as a mechanism for repair of double-strand breaks (DSBs) caused by DNA damage. These DSBs are usually repaired using the sister chromatid of the broken duplex and not the homologous chrom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caudovirales
''Caudoviricetes'' is a class of viruses known as tailed viruses and head-tail viruses (''cauda'' is Latin for "tail"). It is the sole representative of its own phylum, ''Uroviricota'' (from ''ouros'' (ουρος), a Greek word for "tailed" + -viricota). Under the Baltimore classification scheme, the ''Caudoviricetes'' are group I viruses as they have double stranded DNA (dsDNA) genomes, which can be anywhere from 18,000 base pairs to 500,000 base pairs in length. The virus particles have a distinct shape; each virion has an icosahedral head that contains the viral genome, and is attached to a flexible tail by a connector protein. The order encompasses a wide range of viruses, many containing genes of similar nucleotide sequence and function. However, some tailed bacteriophage genomes can vary quite significantly in nucleotide sequence, even among the same genus. Due to their characteristic structure and possession of potentially homologous genes, it is believed these viruse ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacterial Conjugation
Bacterial conjugation is the transfer of genetic material between Bacteria, bacterial cells by direct cell-to-cell contact or by a bridge-like connection between two cells. This takes place through a pilus. It is a parasexual cycle, parasexual mode of reproduction in bacteria. It is a mechanism of horizontal gene transfer as are Transformation (genetics), transformation and Transduction (genetics), transduction although these two other mechanisms do not involve cell-to-cell contact. Classical ''E. coli'' bacterial conjugation is often regarded as the bacterial equivalent of sexual reproduction or mating, since it involves the exchange of genetic material. However, it is not sexual reproduction, since no exchange of gamete occurs, and indeed no biogenesis, generation of a new organism: instead, an existing organism is transformed. During classical ''E. coli'' conjugation, the ''donor'' cell provides a conjugative or mobilizable genetic element that is most often a plasmid or trans ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repressor Protein
In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA. An RNA-binding repressor binds to the mRNA and prevents translation of the mRNA into protein. This blocking or reducing of expression is called repression. Function If an inducer, a molecule that initiates the gene expression, is present, then it can interact with the repressor protein and detach it from the operator. RNA polymerase then can transcribe the message (expressing the gene). A co-repressor is a molecule that can bind to the repressor and make it bind to the operator tightly, which decreases transcription. A repressor that binds with a co-repressor is termed an ''aporepressor'' or ''inactive repressor''. One type of aporepressor is the trp r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
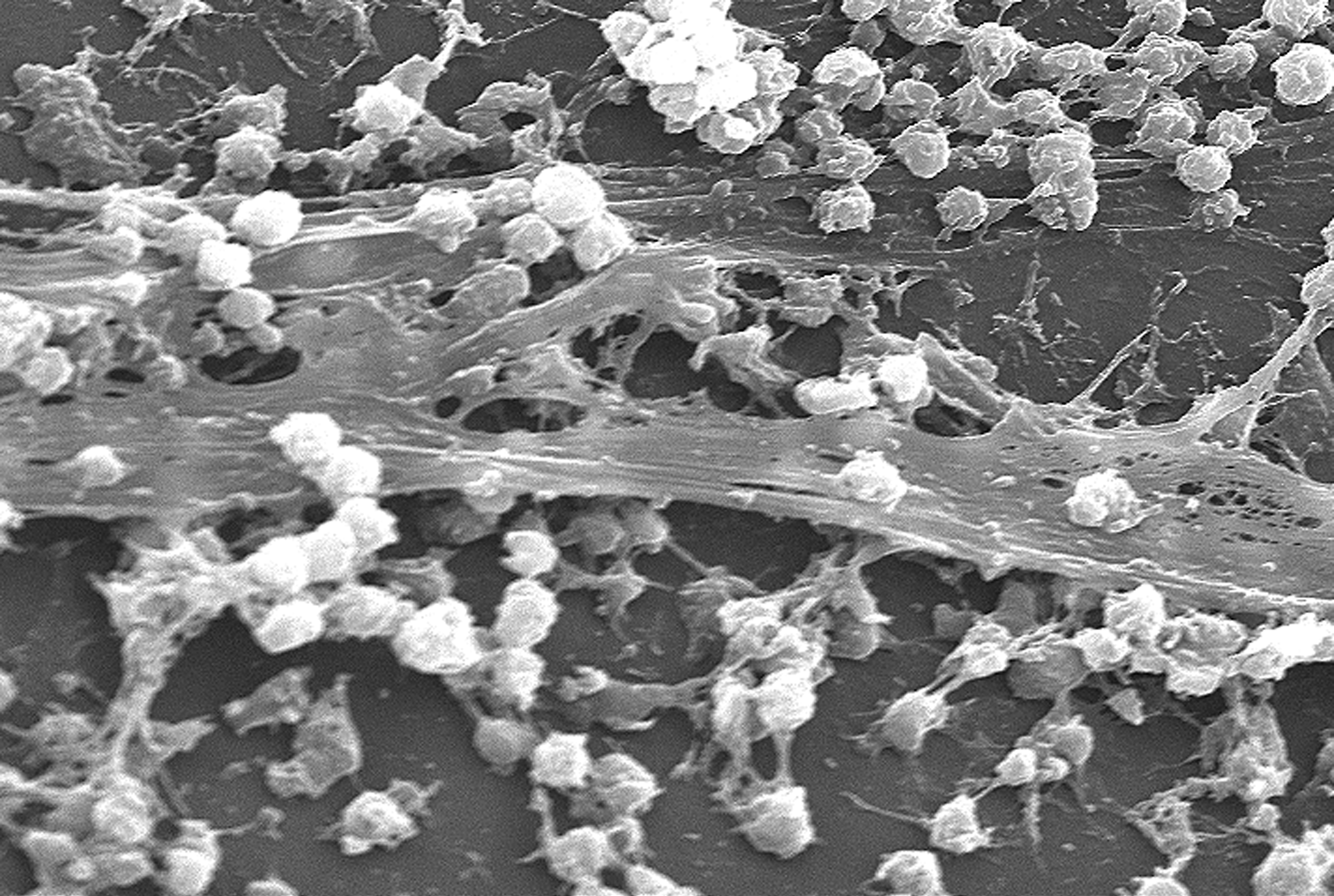
Biofilm
A biofilm is a Syntrophy, syntrophic Microbial consortium, community of microorganisms in which cell (biology), cells cell adhesion, stick to each other and often also to a surface. These adherent cells become embedded within a slimy extracellular matrix that is composed of extracellular polymeric substances (EPSs). The cells within the biofilm produce the EPS components, which are typically a polymeric combination of extracellular polysaccharides, proteins, lipids and DNA. Because they have a three-dimensional structure and represent a community lifestyle for microorganisms, they have been metaphorically described as "cities for microbes". Biofilms may form on living (biotic) or non-living (abiotic) surfaces and can be common in natural, industrial, and hospital settings. They may constitute a microbiome or be a portion of it. The microbial cells growing in a biofilm are physiology, physiologically distinct from planktonic cells of the same organism, which, by contrast, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |