|
Prokaryotic Ubiquitin-like Protein
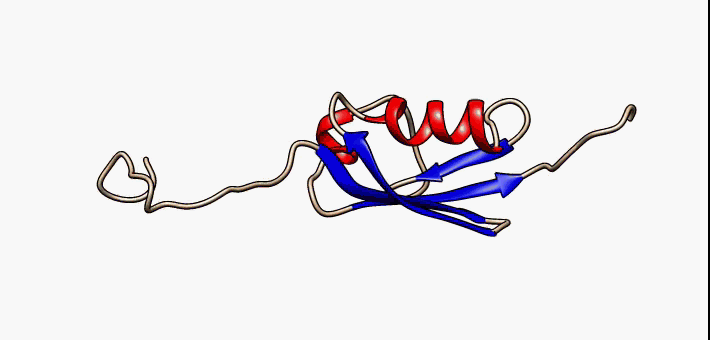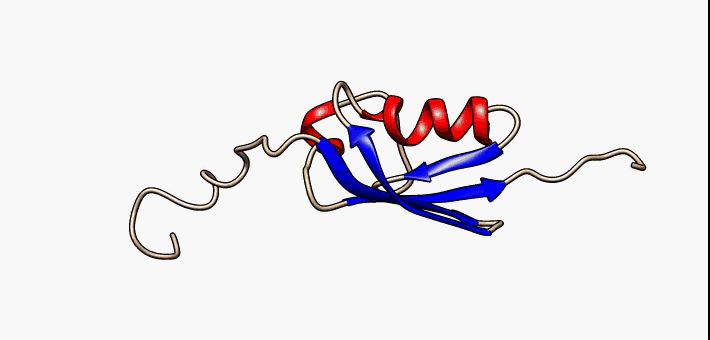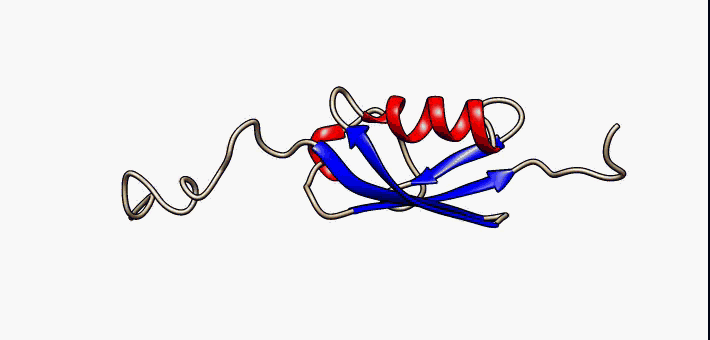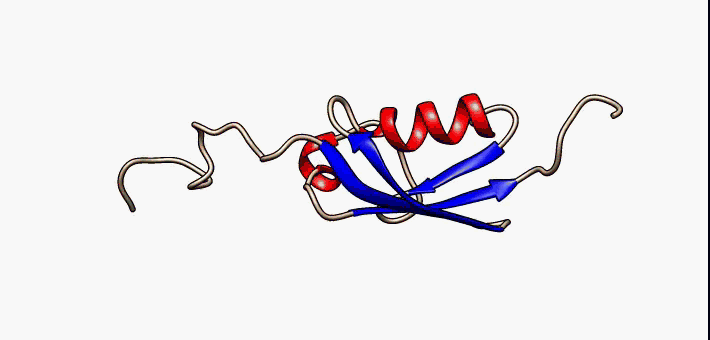
Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin found in the prokaryote ''Mycobacterium tuberculosis''. Like ubiquitin, Pup serves to direct proteins to the proteasome for protein degradation, degradation in the Pup-proteasome system (PPS). However, the enzymology of ubiquitylation and pupylation is different, owing to their distinct evolutionary origins. In contrast to the three-step reaction of ubiquitylation, pupylation requires only two steps, and thus only two enzymes are involved in pupylation. The enzymes involved in pupylation are descended from glutamine synthetase. Similar to ubiquitin, Pup is attached to specific lysine residues of substrate proteins by isopeptide bonds; this is called pupylation. It is then recognized by the protein Mycobacterium proteasomal ATPase (Mpa), in a mechanism that induces folding of Pup. Mpa delivers the substrate protein to the proteasome for protein degradation, degradation by coupling of ATP hydrolysis. The dis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the 26S proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin Bacterial
Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin found in the prokaryote ''Mycobacterium tuberculosis''. Like ubiquitin, Pup serves to direct proteins to the proteasome for degradation in the Pup-proteasome system (PPS). However, the enzymology of ubiquitylation and pupylation is different, owing to their distinct evolutionary origins. In contrast to the three-step reaction of ubiquitylation, pupylation requires only two steps, and thus only two enzymes are involved in pupylation. The enzymes involved in pupylation are descended from glutamine synthetase. Similar to ubiquitin, Pup is attached to specific lysine residues of substrate proteins by isopeptide bonds; this is called pupylation. It is then recognized by the protein Mycobacterium proteasomal ATPase (Mpa), in a mechanism that induces folding of Pup. Mpa delivers the substrate protein to the proteasome for degradation by coupling of ATP hydrolysis. The discovery of Pup indicates that like euka ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nitrospirota
The Nitrospirota are a phylum of bacteria. They include multiple genera such as '' Nitrospira'', the largest. History of knowledge The first member of this phylum, '' Nitrospira marina'', was discovered in 1985. The second member, '' Nitrospira moscoviensis'', was discovered in 1995. '' Nitrospira inopinata'' was discovered in 2015 and cultivated in 2017. Metabolism Nitrospirota contains nitrifying taxa which oxidize nitrite to nitrate (nitrite-oxidizing bacteria, NOB) and commamox bacteria: '' Nitrospira inopinata''. '' Dissulfurispira thermophila'' ( Thermodesulfovibrionia) grows by disproportionation of thiosulfate or elemental sulfur. Phylogeny Taxonomy The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LSPN) and the National Center for Biotechnology Information (NCBI). * Genus ?"''Candidatus'' Magnetomicrobium" Zhang et al. 2021 * Genus ?"''Candidatus'' Thermomagnetovibrio" Lefèvre et al. 2010 * Class Nit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alignment Of UBact From Several Bactrerial Phyla
Alignment may refer to: Archaeology * Alignment (archaeology), a co-linear arrangement of features or structures with external landmarks * Stone alignment, a linear arrangement of upright, parallel megalithic standing stones Biology * Structural alignment, establishing similarities in the 3D structure of protein molecules * Sequence alignment, in bioinformatics, arranging the sequences of DNA, RNA, or protein to identify similarities ** Alignment program, software used in sequence alignment Engineering * Road alignment, the route of a road, defined as a series of horizontal tangents and curves, as defined by planners and surveyors * Railway alignment, three-dimensional geometry of track layouts * Transfer alignment, a process for initializing and calibrating the inertial navigation system on a missile or torpedo * Shaft alignment, in mechanical engineering, aligning two or more shafts with each other * Wheel alignment, automobile wheel and suspension angles which affect p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leptospirillum Ferriphilum
''Leptospirillum ferriphilum'' is an iron-oxidising bacterium able to exist in environments of high acidity, high iron concentrations, and moderate to moderately high temperatures. It is one of the species responsible for the generation of acid mine drainage Acid mine drainage, acid and metalliferous drainage (AMD), or acid rock drainage (ARD) is the outflow of acidic water from metal mines and coal mines. Acid rock drainage occurs naturally within some environments as part of the rock weatherin ... and the principal microbe used in industrial biohydrometallurgy processes to extract metals. Cell morphology ''L. ferriphilum'' is a gram-negative, spiral-shaped bacterium. ''L. ferriphilum'' is an acidophile and a thermotolerant bacteria allowing it to survive in extremely acidic environments and relatively high temperatures. This bacterium is an aerobic organism; it can only survive and grow in an oxygenated environment. Phylogeny ''Leptospirillum ferriphilum'' is from t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intrinsically Disordered Proteins
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered protein tertiary structure, three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like Protein aggregation, aggregates, or flexible linkers in large multi-Protein domain, domain proteins. They are sometimes considered as a separate class of proteins along with globular protein, globular, fibrous protein, fibrous and membrane proteins. IDPs are a very large and functionally important class of proteins. They are most numerous in Eukaryote, eukaryotes, with an estimated 30-40% of residues in the eukaryotic proteome located in disordered regions. Disorder is present in around 70% of proteins, either in the form of disordered tails or flexible linkers. Proteins can also be entirely disordered and lack a define ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of life forms alongside the two groups of prokaryotes: the Bacteria and the Archaea. Eukaryotes represent a small minority of the number of organisms, but given their generally much larger size, their collective global biomass is much larger than that of prokaryotes. The eukaryotes emerged within the archaeal kingdom Promethearchaeati and its sole phylum Promethearchaeota. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among the Archaea. Eukaryotes first emerged during the Paleoproterozoic, likely as flagellated cells. The leading evolutionary theory is they were created by symbiogenesis between an anaerobic Promethearchaeati archaean and an aerobic proteobacterium, which form ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the 26S proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Israel
Israel, officially the State of Israel, is a country in West Asia. It Borders of Israel, shares borders with Lebanon to the north, Syria to the north-east, Jordan to the east, Egypt to the south-west, and the Mediterranean Sea to the west. Israeli-occupied territories, It occupies the Occupied Palestinian territories, Palestinian territories of the West Bank in the east and the Gaza Strip in the south-west. Israel also has a small coastline on the Red Sea at its southernmost point, and part of the Dead Sea lies along its eastern border. Status of Jerusalem, Its proclaimed capital is Jerusalem, while Tel Aviv is the country's Gush Dan, largest urban area and Economy of Israel, economic center. Israel is located in a region known as the Land of Israel, synonymous with the Palestine (region), Palestine region, the Holy Land, and Canaan. In antiquity, it was home to the Canaanite civilisation followed by the History of ancient Israel and Judah, kingdoms of Israel and Judah. Situate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aaron Ciechanover
Aaron Ciechanover ( ; ; born October 1, 1947) is an Israeli biologist who won the Nobel Prize in Chemistry for characterizing the method that cells use to degrade and recycle proteins using ubiquitin. Biography Early life Ciechanover was born in Haifa, British Mandate of Palestine on 1 October 1947 into a Jewish family. He is the son of Bluma (Lubashevsky), a teacher of English, and Yitzhak Ciechanover, an office worker in a law firm. His parents immigrated to Israel from Poland in the 1920s. Education He earned a master's degree in science in 1971 and graduated from Hadassah Medical School in Jerusalem in 1974. He received his doctorate in biochemistry in 1981 from the Technion – Israel Institute of Technology in Haifa before conducting postdoctoral research in the laboratory of Harvey Lodish at the Whitehead Institute at MIT from 1981 to 1984. Recent Ciechanover is currently a Technion Distinguished Research Professor in the Ruth and Bruce Rappaport Faculty of Med ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |