|
Parthanatos
Parthanatos (derived from the Greek Θάνατος, "Death") is a form of programmed cell death that is distinct from other cell death processes such as necrosis and apoptosis. While necrosis is caused by acute cell injury resulting in traumatic cell death and apoptosis is a highly controlled process signalled by apoptotic intracellular signals, parthanatos is caused by the accumulation of Poly (ADP ribose) (PAR) and the nuclear translocation of apoptosis-inducing factor (AIF) from mitochondria. Parthanatos is also known as PARP-1 dependent cell death. PARP-1 mediates parthanatos when it is over-activated in response to extreme genomic stress and synthesizes PAR which causes nuclear translocation of AIF. Parthanatos is involved in diseases that afflict hundreds of millions of people worldwide. Well known diseases involving parthanatos include Parkinson's disease, stroke, heart attack, and diabetes. It also has potential use as a treatment for ameliorating disease and various m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Programmed Cell Death
Programmed cell death (PCD; sometimes referred to as cellular suicide) is the death of a cell (biology), cell as a result of events inside of a cell, such as apoptosis or autophagy. PCD is carried out in a biological process, which usually confers advantage during an organism's biological life cycle, lifecycle. For example, the Limb development, differentiation of fingers and toes in a developing human embryo occurs because cells between the fingers apoptose; the result is that the digits are separate. PCD serves fundamental functions during both plant and animal tissue development. Apoptosis and autophagy are both forms of programmed cell death. Necrosis is the death of a cell caused by external factors such as trauma or infection and occurs in several different forms. Necrosis was long seen as a non-physiological process that occurs as a result of infection or injury, but in the 2000s, a form of programmed necrosis, called necroptosis, was recognized as an alternative form of pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apoptosis-inducing Factor
Apoptosis inducing factor is involved in initiating a caspase-independent pathway of apoptosis (positive intrinsic regulator of apoptosis) by causing DNA fragmentation and chromatin condensation. Apoptosis inducing factor is a flavoprotein. It also acts as an NADH oxidase. Another AIF function is to regulate the permeability of the mitochondrial membrane upon apoptosis. Normally it is found behind the outer membrane of the mitochondrion and is therefore secluded from the nucleus. However, when the mitochondrion is damaged, it moves to the cytosol and to the nucleus. Inactivation of AIF leads to resistance of embryonic stem cells to death following the withdrawal of growth factors indicating that it is involved in apoptosis. Function Apoptosis Inducing Factor (AIF) is a protein that triggers chromatin condensation and DNA fragmentation in a cell in order to induce programmed cell death. The mitochondrial AIF protein was found to be a caspase-independent death effector that ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PARP1
Poly DP-ribosepolymerase 1 (PARP-1) also known as NAD+ ADP-ribosyltransferase 1 or poly DP-ribosesynthase 1 is an enzyme that in humans is encoded by the ''PARP1'' gene. It is the most abundant of the PARP family of enzymes, accounting for 90% of the NAD+ used by the family. PARP1 is mostly present in cell nucleus, but cytosolic fraction of this protein was also reported. Function PARP1 works: * By using NAD+ to synthesize poly ADP ribose (PAR) and transferring PAR moieties to proteins ( ADP-ribosylation). * In conjunction with BRCA, which acts on double strands; members of the PARP family act on single strands; or, when BRCA fails, PARP takes over those jobs as well (in a DNA repair context). PARP1 is involved in: * Differentiation, proliferation, and tumor transformation * Normal or abnormal recovery from DNA damage * May be the site of mutation in Fanconi anemia * Induction of inflammation. * The pathophysiology of type I diabetes. PARP1 is activated by: * Helico ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
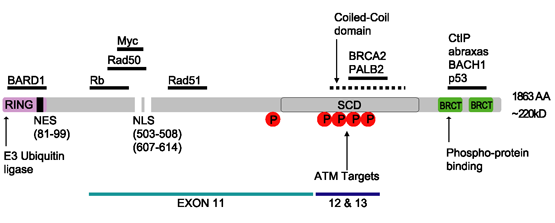
BRCA1
Breast cancer type 1 susceptibility protein is a protein that in humans is encoded by the ''BRCA1'' () gene. Orthologs are common in other vertebrate species, whereas invertebrate genomes may encode a more distantly related gene. ''BRCA1'' is a human tumor suppressor gene (also known as a caretaker gene) and is responsible for repairing DNA. ''BRCA1'' and ''BRCA2'' are unrelated proteins, but both are normally expressed in the cells of breast and other tissue, where they help repair damaged DNA, or destroy cells if DNA cannot be repaired. They are involved in the repair of chromosomal damage with an important role in the error-free repair of DNA double-strand breaks. If ''BRCA1'' or ''BRCA2'' itself is damaged by a BRCA mutation, damaged DNA is not repaired properly, and this increases the risk for breast cancer. ''BRCA1'' and ''BRCA2'' have been described as "breast cancer susceptibility genes" and "breast cancer susceptibility proteins". The predominant allele has a normal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
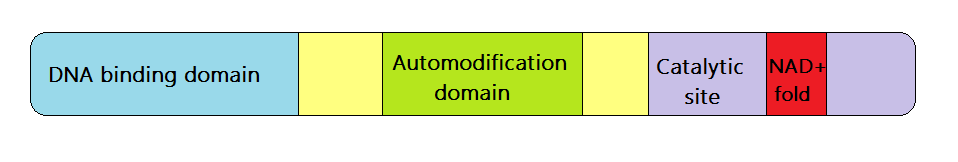
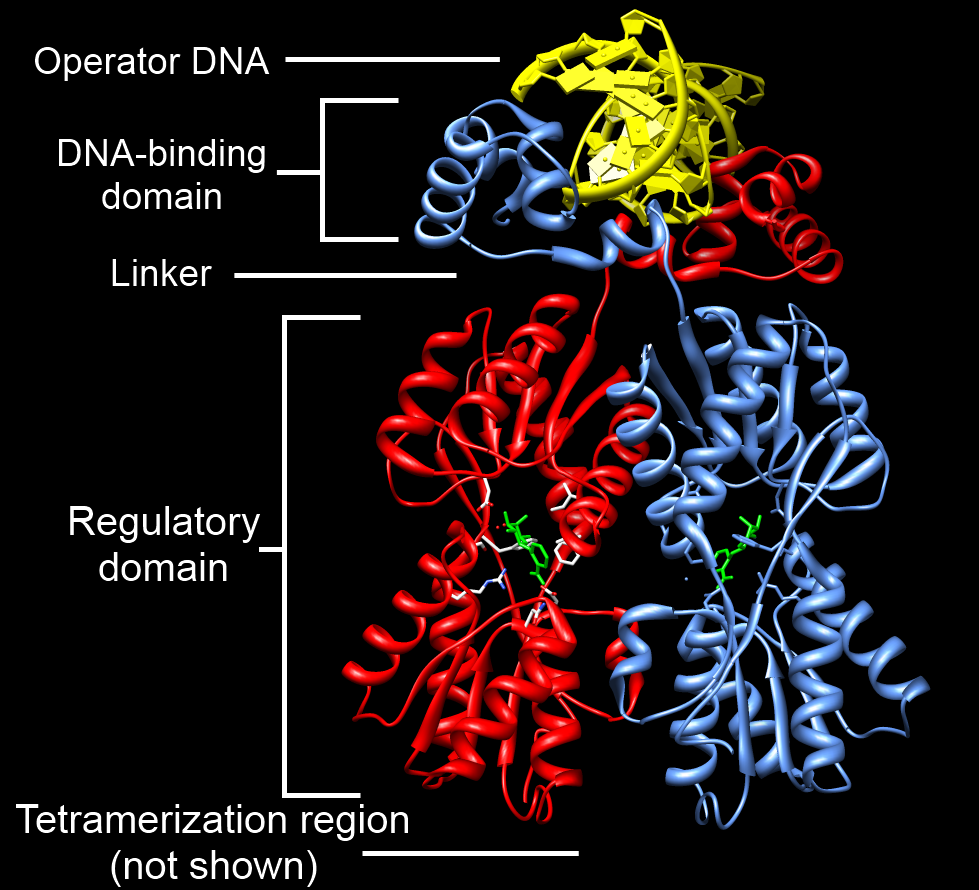
DNA-binding Domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. Function One or more DNA-binding domains are often part of a larger protein consisting of further protein domains with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping. DNA-binding domains with functions involving DNA structure have biological roles in DNA replication, repair, storage, and modification, such as methylation. Many proteins involved in the regulation of gene expression contain DNA-binding domains. For example, proteins that regulate transcription ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalytic Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) and residues that catalyse a reaction of that substrate (catalytic site). Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their function. The active sit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hyperglycemia
Hyperglycemia is a condition in which an excessive amount of glucose circulates in the blood plasma. This is generally a blood sugar level higher than 11.1 mmol/L (200 mg/dL), but symptoms may not start to become noticeable until even higher values such as 13.9–16.7 mmol/L (~250–300 mg/dL). A subject with a consistent range between ~5.6 and ~7 mmol/L (100–126 mg/dL) ( American Diabetes Association guidelines) is considered slightly hyperglycemic, and above 7 mmol/L (126 mg/dL) is generally held to have diabetes. For diabetics, glucose levels that are considered to be too hyperglycemic can vary from person to person, mainly due to the person's renal threshold of glucose and overall glucose tolerance. On average, however, chronic levels above 10–12 mmol/L (180–216 mg/dL) can produce noticeable organ damage over time. Signs and symptoms The degree of hyperglycemia can change over time depending on the metabolic cause, for example, impaired ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ADP-ribose
Adenosine diphosphate ribose (ADPR) is an ester molecule formed into chains by the enzyme poly ADP ribose polymerase. ADPR is created from cyclic ADP-ribose (cADPR) by the CD38 enzyme using nicotinamide adenine dinucleotide (NAD+) as a cofactor. ADPR binds to and activates the TRPM2 ion channel. ADPR is the most potent agonist of the TRPM2 channel. cADPR also binds to TPRM2, and the action of both molecules is synergistic, with both molecules enhancing the action of the other molecule in activating the TRPM2 channel. See also * Adenosine diphosphate * ADP-ribosylation * Ribose Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally-occurring form, , is a component of the ribonucleotides from which RNA is built, and so this compou ... References Nucleotides Organophosphates NADH dehydrogenase inhibitors Phosphate esters {{biochemistry-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). E ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |