|
PKNOX2
PBX/Knotted 1 Homeobox 2 (PKNOX2) protein belongs to the three amino acid loop extension (TALE) class of homeodomain proteins, and is encoded by ''PKNOX2'' gene in humans. The protein regulates the transcription of other genes and affects anatomical development. Function PKNOX2 protein regulates expression of other genes by binding to DNA in a sequence-specific manner, i.e. to specific DNA sequences. It binds to unique DNA recognition motifs, acting as a nuclear transcription factor. The nuclear transcription factor is a protein that regulates transcription of genetic information from DNA to messenger RNA. Other functions of PKNOX2 include actin filament binding. An important paralog of the ''PKNOX2'' gene is ''PKNOX1''. Structure PKNOX2 belongs to the three amino acid loop extension (TALE) class of homeodomain proteins. This class is characterized by a 3-amino acid extension between alpha helix 1 and 2 in the homeodomain. Homeodomain proteins are sequence-specific transc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PKNOX1
PBX/Knotted 1 Homeobox 1 (PKNOX1) is a protein that in humans is encoded by the ''PKNOX1'' gene. An important paralog of this gene is '' PKNOX2''. Function PKNOX1 belongs to the three amino acid loop extension (TALE) class of homeodomain transcription factors that form transcriptionally active complexes involved in development and organogenesis. PKNOX1 is essential for embryogenesis, but it can also act as a tumor suppressor in adulthood An adult is an animal that has reached full growth. The biological definition of the word means an animal reaching sexual maturity and thus capable of reproduction. In the human context, the term ''adult'' has meanings associated with social an .... References Further reading * * * * * * * * * * * * * * * * External links * Transcription factors {{gene-21-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homeobox
A homeobox is a Nucleic acid sequence, DNA sequence, around 180 base pairs long, that regulates large-scale anatomical features in the early stages of embryonic development. Mutations in a homeobox may change large-scale anatomical features of the full-grown organism. Homeoboxes are found within genes that are involved in the regulation of patterns of anatomical development (morphogenesis) in animals, fungus, fungi, plants, and numerous single cell eukaryotes. Homeobox genes encode homeodomain protein products that are transcription factors sharing a characteristic protein fold structure that binds DNA to regulate expression of target genes. Homeodomain proteins regulate gene expression and cell differentiation during early embryonic development, thus mutations in homeobox genes can cause developmental disorders. Homeosis is a term coined by William Bateson to describe the outright replacement of a discrete body part with another body part, e.g. antennapedia—replacement of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-binding Protein
DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. Examples DNA-binding proteins include transcription factors which modulate the process of transcription, various polymerases, nucleases which cleave DNA molecules, and histones which are involved in chromosome packaging and transcription in the cell nucleus. DNA-binding proteins can incorporate such domains as the zinc finger, the helix-turn-helix, and the leucine zipper (among many others) that facilitate binding to nucleic acid. There are also more unusual examples such as transcription activator like effectors. Non-specific DNA-protein interactions Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ''Asn, followed by anything but Pro, followed by either Ser or Thr, followed by anything but Pro residue''. Overview When a sequence motif appears in the exon of a gene, it may encode the " structural motif" of a protein; that is a stereotypical element of the overall structure of the protein. Nevertheless, motifs need not be associated with a distinctive secondary structure. " Noncoding" sequences are not translated into proteins, and nucleic acids with such motifs need not deviate from the typical shape (e.g. the "B-form" DNA double helix). Outside of gene exons, there exist regulatory sequence motifs and motifs within the " junk", such as satellite DNA. Some of these are believed to affect the shape of nucleic acids (see for example ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are Gene expression, expressed in the desired Cell (biology), cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are approximately 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transcribed into RNA molecules called non-coding RNAs (ncRNAs). Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a Complementarity (molecular biology), complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, Antiparallel (biochemistry), antiparallel RNA strand called a primary transcript. In virology, the term transcription is used when referring to mRNA synthesis from a viral RNA molecule. The genome of many Orthornavirae, RNA viruses is composed of Sense (molecular biology), negative-sense RNA which acts as a template for positive sense viral messenger RNA - a necessary step in the synthesis of viral proteins needed for viral replication. This process ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and the ribosome creates the protein utilizing amino acids carried by transfer RNA (tRNA). This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, genetic inf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Actin
Actin is a family of globular multi-functional proteins that form microfilaments in the cytoskeleton, and the thin filaments in muscle fibrils. It is found in essentially all eukaryotic cells, where it may be present at a concentration of over 100 μM; its mass is roughly 42 kDa, with a diameter of 4 to 7 nm. An actin protein is the monomeric subunit of two types of filaments in cells: microfilaments, one of the three major components of the cytoskeleton, and thin filaments, part of the contractile apparatus in muscle cells. It can be present as either a free monomer called G-actin (globular) or as part of a linear polymer microfilament called F-actin (filamentous), both of which are essential for such important cellular functions as the mobility and contraction of cells during cell division. Actin participates in many important cellular processes, including muscle contraction, cell motility, cell division and cytokinesis, vesicle and organelle mov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paralog
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
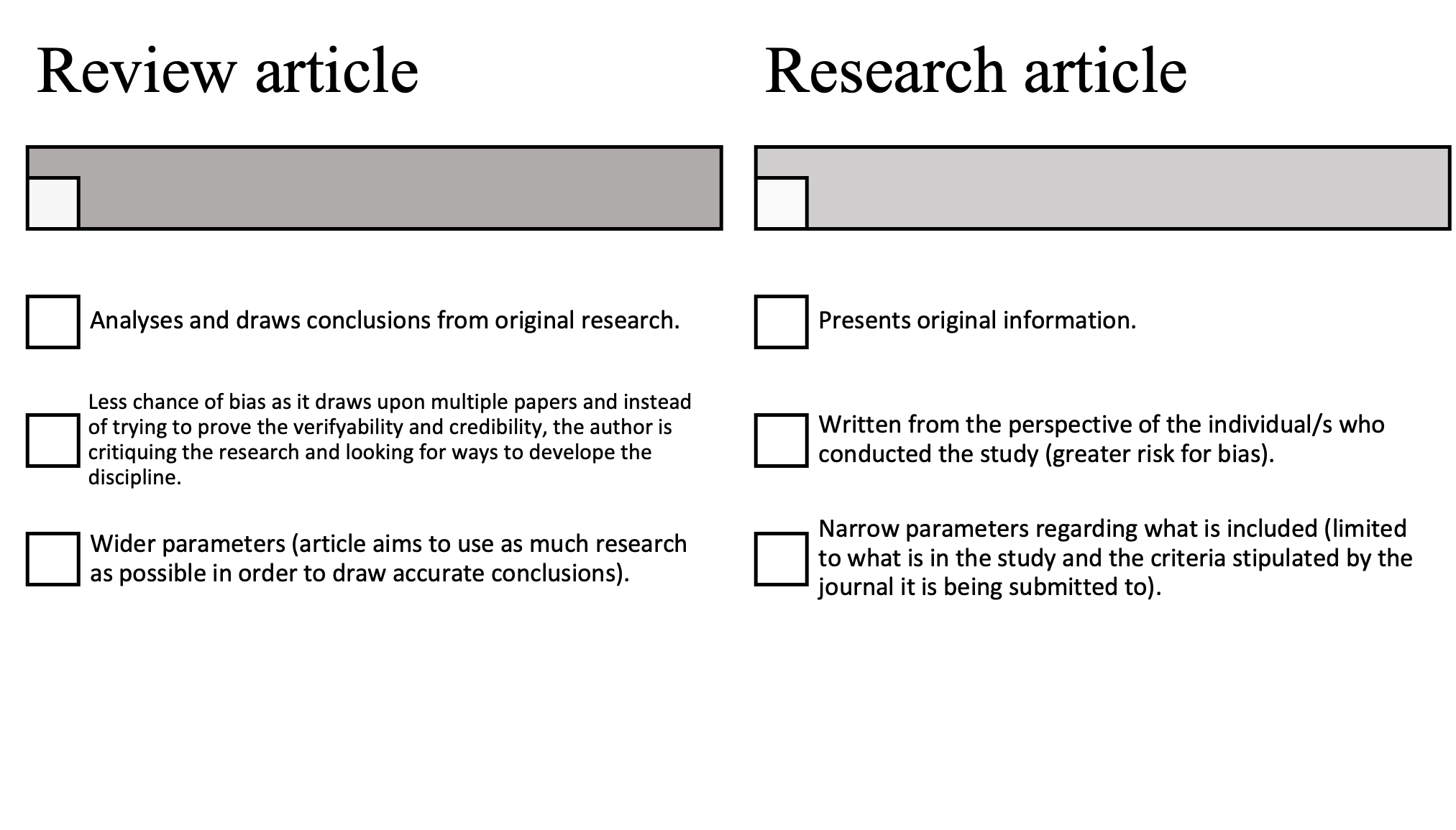
Review Article
A review article is an article (publishing), article that summarizes the current Status quaestionis, state of understanding on a topic within a certain discipline. A review article is generally considered a secondary source since it may analyze and discuss the method and conclusions in previously published studies. It resembles a survey article or, in news publishing, overview article, which also surveys and summarizes previously published primary source, primary and secondary sources, instead of reporting new facts and results. Survey articles are however considered tertiary sources, since they do not provide additional analysis and synthesis of new conclusions. A review of such sources is often referred to as a tertiary review. Academic publications that specialize in review articles are known as review journals. Review journals have their own requirements for the review articles they accept, so review articles may vary slightly depending on the journal they are being submitte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
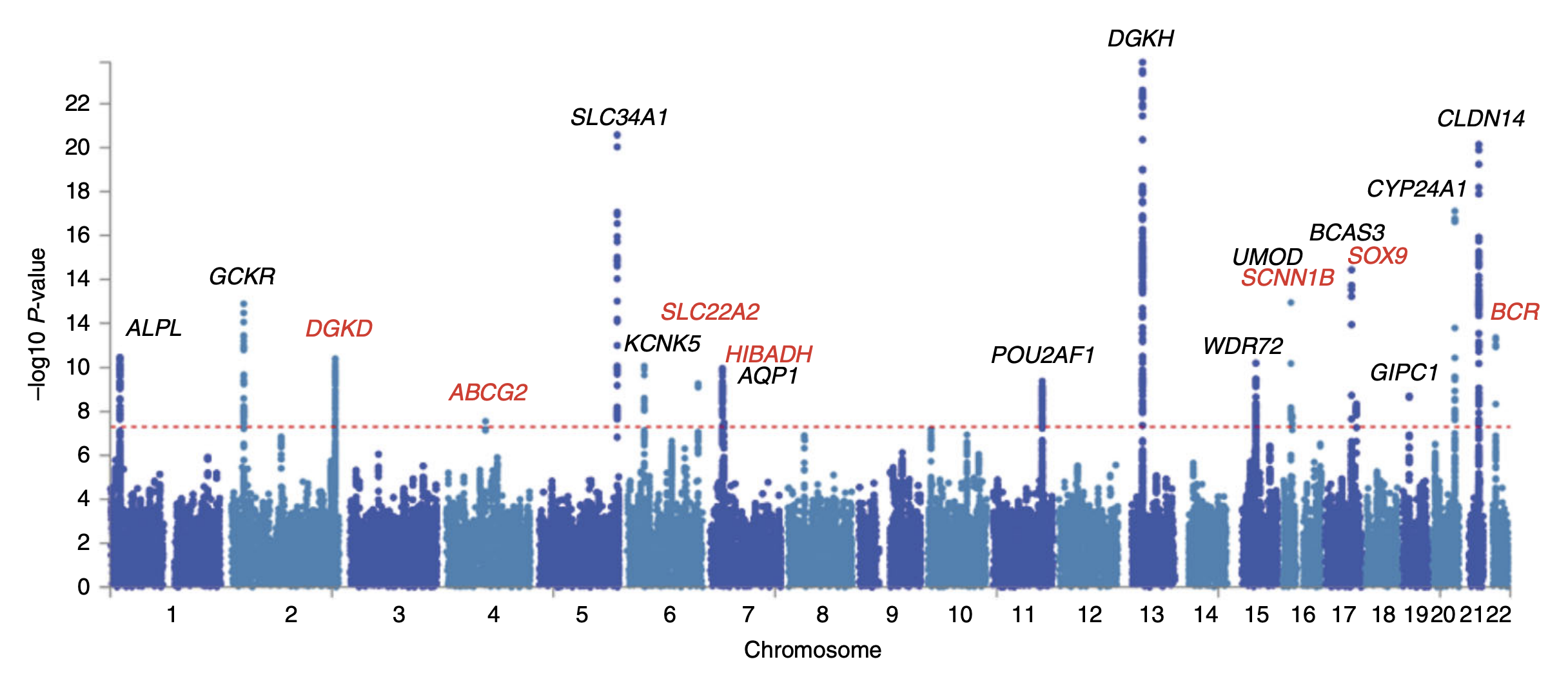
Genome-wide Association Study
In genomics, a genome-wide association study (GWA study, or GWAS), is an observational study of a genome-wide set of Single-nucleotide polymorphism, genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms. When applied to human data, GWA studies compare the DNA of participants having varying phenotypes for a particular trait or disease. These participants may be people with a disease (cases) and similar people without the disease (controls), or they may be people with different phenotypes for a particular trait, for example blood pressure. This approach is known as phenotype-first, in which the participants are classified first by their clinical manifestation(s), as opposed to Genotype-first approach, genotype-first. Each person gives a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |