|
PER10129 01
The PER1 gene encodes the period circadian protein homolog 1 protein in humans. Function The PER1 protein is important to the maintenance of circadian rhythms in cells, and may also play a role in the development of cancer. This gene is a member of the period family of genes. It is expressed with a daily oscillating circadian rhythm, or an oscillation that cycles with a period of approximately 24 hours. PER1 is most notably expressed in the region of the brain called the suprachiasmatic nucleus (SCN), which is the primary circadian pacemaker in the mammalian brain. PER1 is also expressed throughout mammalian peripheral tissues. Genes in this family encode components of the circadian rhythms of locomotor activity, metabolism, and behavior. Circadian expression of PER1 in the suprachiasmatic nucleus will free-run in constant darkness, meaning that the 24-hour period of the cycle will persist without the aid of external light cues. Subsequently, a shift in the light/dark cycle ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CRY1
Cryptochromes (from the Greek κρυπτός χρώμα, "hidden colour") are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields in a number of species. The name ''cryptochrome'' was proposed as a '' portmanteau'' combining the ''chromatic'' nature of the photoreceptor, and the '' cryptogamic'' organisms on which many blue-light studies were carried out. The two genes ''Cry1'' and ''Cry2'' code the two cryptochrome proteins CRY1 and CRY2. In insects and plants, CRY1 regulates the circadian clock in a light-dependent fashion, whereas in mammals, CRY1 and CRY2 act as light-independent inhibitors of CLOCK- BMAL1 components of the circadian clock. In plants, blue-light photoreception can be used to cue developmental signals. Besides chlorophylls, cryptochromes are the only proteins known to form photoinduced radical-pairs '' in vivo''. These appear to enable s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Suprachiasmatic Nucleus
The suprachiasmatic nucleus or nuclei (SCN) is a tiny region of the brain in the hypothalamus, situated directly above the optic chiasm. It is responsible for controlling circadian rhythms. The neuronal and hormonal activities it generates regulate many different body functions in a 24-hour cycle. The mouse SCN contains approximately 20,000 neurons. The SCN interacts with many other regions of the brain. It contains several cell types and several different peptides (including vasopressin and vasoactive intestinal peptide) and neurotransmitters. Neuroanatomy The SCN is situated in the anterior part of the hypothalamus immediately dorsal, or ''superior'' (hence supra) to the optic chiasm (CHO) bilateral to (on either side of) the third ventricle. The nucleus can be divided into ventrolateral and dorsolateral portions, also known as the core and shell, respectively. These regions differ in their expression of the clock genes, the core expresses them in response to stimuli w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ortholog
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties (' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PER3
The PER3 gene encodes the period circadian protein homolog 3 protein in humans. PER3 is a paralog to the PER1 and PER2 genes. It is a circadian gene associate with delayed sleep phase syndrome in humans. History The Per3 gene was independently cloned by two research groups (Kobe University School of Medicine and the Harvard Medical School) who both published their discovery in June 1998. The mammalian Per3 was discovered by searching for homologous cDNA sequences to Per2. The amino acid sequence of the mouse PERIOD3 protein (mPER3) is between 37-56% similar to the other two PER proteins. Function This gene is a member of the Period family of genes. It is expressed in a circadian pattern in the suprachiasmatic nucleus (SCN), the primary circadian pacemaker in the mammalian brain. Genes in this family encode components of the circadian rhythms of locomotor activity, metabolism, and behavior. Circadian expression in the SCN continues in constant darkness, and a shift in the l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology (biology)
In biology, homology is similarity due to shared ancestry between a pair of structures or genes in different taxa. A common example of homologous structures is the forelimbs of vertebrates, where the wings of bats and birds, the arms of primates, the front flippers of whales and the forelegs of four-legged vertebrates like dogs and crocodiles are all derived from the same ancestral tetrapod structure. Evolutionary biology explains homologous structures adapted to different purposes as the result of descent with modification from a common ancestor. The term was first applied to biology in a non-evolutionary context by the anatomist Richard Owen in 1843. Homology was later explained by Charles Darwin's theory of evolution in 1859, but had been observed before this, from Aristotle onwards, and it was explicitly analysed by Pierre Belon in 1555. In developmental biology, organs that developed in the embryo in the same manner and from similar origins, such as from matc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CHEK2
CHEK2 (Checkpoint kinase 2) is a tumor suppressor gene that encodes the protein CHK2, a serine-threonine kinase. CHK2 is involved in DNA repair, cell cycle arrest or apoptosis in response to DNA damage. Mutations to the CHEK2 gene have been linked to a wide range of cancers. Gene location The CHEK2 gene is located on the long (q) arm of chromosome 22 at position 12.1. Its location on chromosome 22 stretches from base pair 28,687,742 to base pair 28,741,904. Protein structure The CHEK2 protein encoded by the CHEK2 gene is a serine threonine kinase. The protein consists of 543 amino acids and the following domains: * N-terminal SQ/TQ cluster domain (SCD) * Central forkhead-associated (FHA) domain * C-terminal serine/threonine kinase domain (KD) The SCD domain contains multiple SQ/TQ motifs that serve as sites for phosphorylation in response to DNA damage. The most notable and frequently phosphorylated site being Thr68. CHK2 appears as a monomer in its inactive state. H ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
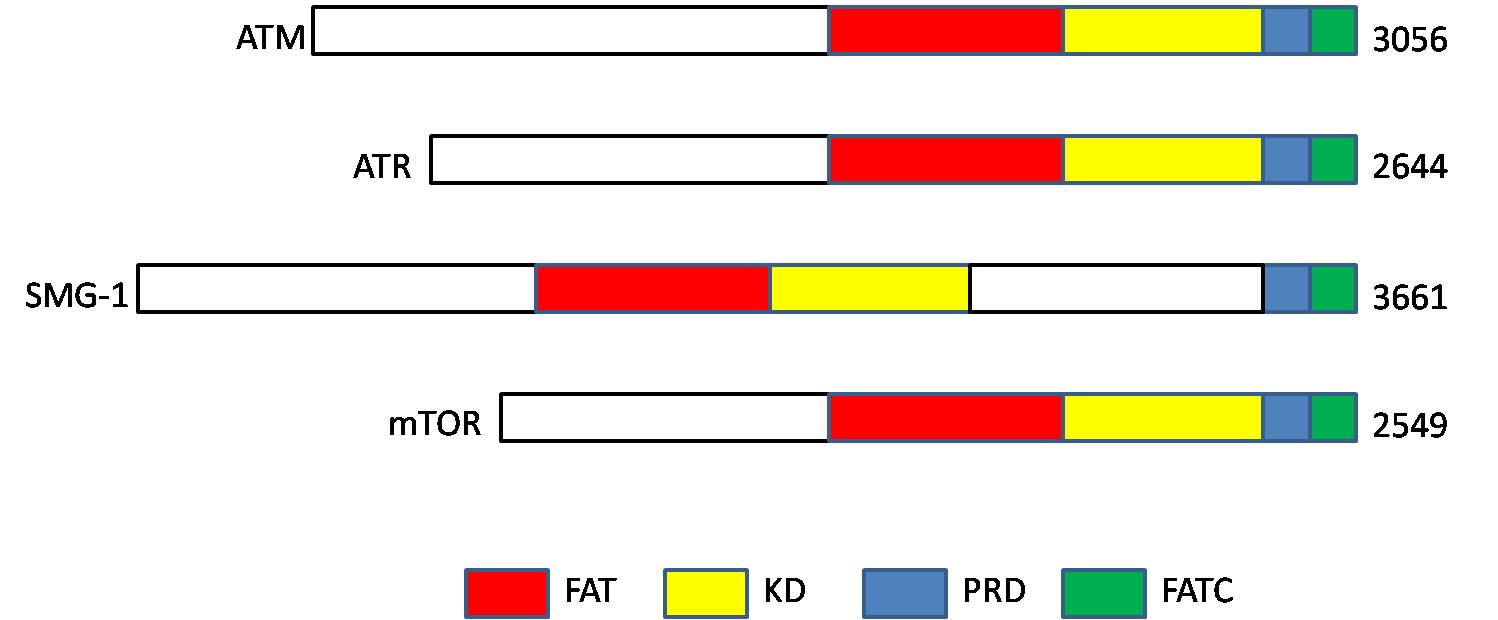
Ataxia Telangiectasia Mutated
ATM serine/threonine kinase or Ataxia-telangiectasia mutated, symbol ATM, is a serine/threonine protein kinase that is recruited and activated by DNA double-strand breaks. It phosphorylates several key proteins that initiate activation of the DNA damage checkpoint, leading to cell cycle arrest, DNA repair or apoptosis. Several of these targets, including p53, CHK2, BRCA1, NBS1 and H2AX are tumor suppressors. In 1995, the gene was discovered by Yosef Shiloh who named its product ATM since he found that its mutations are responsible for the disorder ataxia–telangiectasia. In 1998, the Shiloh and Kastan laboratories independently showed that ATM is a protein kinase whose activity is enhanced by DNA damage. Introduction Throughout the cell cycle DNA is monitored for damage. Damages result from errors during replication, by-products of metabolism, general toxic drugs or ionizing radiation. The cell cycle has different DNA damage checkpoints, which inhibit the next or maintain ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Localization Sequence
A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. An NLS has the opposite function of a nuclear export signal (NES), which targets proteins out of the nucleus. Types Classical These types of NLSs can be further classified as either monopartite or bipartite. The major structural differences between the two are that the two basic amino acid clusters in bipartite NLSs are separated by a relatively short spacer sequence (hence bipartite - 2 parts), while monopartite NLSs are not. The first NLS to be discovered was the sequence PKKKRKV in the SV40 Large T-antigen (a monopartite NLS). The NLS of nucleoplasmin, KR AATKKAGQAKKK, is the prototype of the ubiquitous bipartite signal: two clu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PAS Domain
A Per-Arnt-Sim (PAS) domain is a protein domain found in all kingdoms of life. Generally, the PAS domain acts as a molecular sensor, whereby small molecules and other proteins associate via binding of the PAS domain. Due to this sensing capability, the PAS domain has been shown as the key structural motif involved in protein-protein interactions of the circadian clock, and it is also a common motif found in signaling proteins, where it functions as a signaling sensor. Discovery PAS domains are found in a large number of organisms from bacteria to mammals. The PAS domain was named after the three proteins in which it was first discovered: * Per – period circadian protein * Arnt – aryl hydrocarbon receptor nuclear translocator protein * Sim – single-minded protein Since the initial discovery of the PAS domain, a large quantity of PAS domain binding sites have been discovered in bacteria and eukaryotes. A subset called PAS LOV proteins are responsive to oxygen, light a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CRY2
Cryptochromes (from the Greek κρυπτός χρώμα, "hidden colour") are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields in a number of species. The name ''cryptochrome'' was proposed as a ''portmanteau'' combining the ''chromatic'' nature of the photoreceptor, and the '' cryptogamic'' organisms on which many blue-light studies were carried out. The two genes ''Cry1'' and ''Cry2'' code the two cryptochrome proteins CRY1 and CRY2. In insects and plants, CRY1 regulates the circadian clock in a light-dependent fashion, whereas in mammals, CRY1 and CRY2 act as light-independent inhibitors of CLOCK- BMAL1 components of the circadian clock. In plants, blue-light photoreception can be used to cue developmental signals. Besides chlorophylls, cryptochromes are the only proteins known to form photoinduced radical-pairs ''in vivo''. These appear to enable some anima ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
E-box
An E-box (enhancer box) is a DNA response element found in some eukaryotes that acts as a protein-binding site and has been found to regulate gene expression in neurons, muscles, and other tissues. Its specific DNA sequence, CANNTG (where N can be any nucleotide), with a palindromic canonical sequence of CACGTG, is recognized and bound by transcription factors to initiate gene transcription. Once the transcription factors bind to the promoters through the E-box, other enzymes can bind to the promoter and facilitate transcription from DNA to mRNA. Discovery The E-box was discovered in a collaboration between Susumu Tonegawa's and Walter Gilbert's laboratories in 1985 as a control element in immunoglobulin heavy-chain enhancer. They found that a region of 140 base pairs in the tissue-specific transcriptional enhancer element was sufficient for different levels of transcription enhancement in different tissues and sequences. They suggested that proteins made by specific tissues act ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |