|
Ornithine Translocase
Ornithine translocase is responsible for transporting ornithine from the cytosol into the mitochondria in the urea cycle. It is highly expressed in the liver and pancreas. Pathology A disorder is associated with ornithine translocase deficiency, and a form of hyperammonemia. See also * Translocase Translocase is a general term for a protein that assists in moving another molecule, usually across a cell membrane. These Enzyme, enzymes catalyze the movement of ions or molecules across membranes or their separation within membranes. The reactio ... External links * * * Solute carrier family Urea cycle {{membrane-protein-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Solute Carrier Family
The solute carrier (SLC) group of membrane transport proteins include over 400 members organized into 66 families. Most members of the SLC group are located in the cell membrane. The SLC gene nomenclature system was originally proposed by the HUGO Gene Nomenclature Committee (HGNC) and is the basis for the official HGNC names of the genes that encode these transporters. A more general transmembrane transporter classification can be found in TCDB, TCDB database. Solutes that are transported by the various SLC group members are extremely diverse and include both charged and uncharged organic molecules as well as inorganic ions and the gas Ammonia transporter, ammonia. As is typical of integral membrane proteins, SLCs contain a number of hydrophobic transmembrane Alpha helix, alpha helices connected to each other by hydrophilic intra- and extra-cellular loops. Depending on the SLC, these transporters are functional as either monomers or obligate homo- or hetero-oligomers. Many SLC fam ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondrial Carrier
Mitochondrial carriers are proteins from solute carrier family 25 which transfer molecules across the membranes of the mitochondria. Mitochondrial carriers are also classified in thTransporter Classification Database The Mitochondrial Carrier (MC) Superfamily has been expanded to include both the original Mitochondrial Carrier (MC) familyTC# 2.A.29 and the Mitochondrial Inner/Outer Membrane Fusion (MMF) familyTC# 1.N.6. Phylogeny Members of the MC family (SLC25)TC# 2.A.29 are found exclusively in eukaryotic organelles although they are nuclearly encoded. Most are found in mitochondria, but some are found in peroxisomes of animals, in hydrogenosomes of anaerobic fungi, and in amyloplasts of plants. SLC25 is the largest solute transporter family in humans. 53 members have been identified in human genome, 58 in '' A. thaliana'' and 35 in '' S. cerevisiae''. The functions of approximately 30% of the human SLC25 proteins are unknown, but most of the yeast homologues have been fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ornithine
Ornithine is a non-proteinogenic α-amino acid that plays a role in the urea cycle. It is not incorporated into proteins during translation. Ornithine is abnormally accumulated in the body in ornithine transcarbamylase deficiency, a disorder of the urea cycle. The Moiety (chemistry), moiety derived from ornithine is called ornithyl. Role in urea cycle L-Ornithine is one of the products of the action of the enzyme arginase on L-arginine, creating urea. Therefore, ornithine is a central component of the urea cycle, which enables the disposal of excess nitrogen. Ornithine itself is recycled and, in a sense, acts as a catalyst. First, ammonia is converted into carbamoyl phosphate () by carbamoyl phosphate synthetase. Ornithine transcarbamylase then catalyzes the reaction between carbamoyl phosphate and ornithine to form citrulline and phosphate (Pi). Another amino group is contributed by aspartate, leading to the formation of arginine and the byproduct fumarate. The resulting arginine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
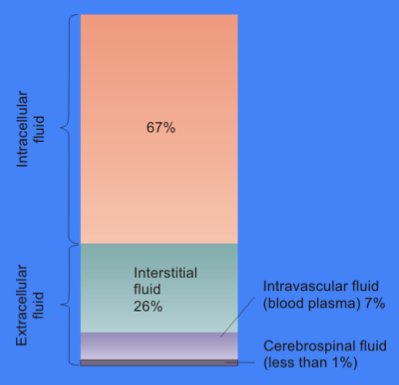
Cytosol
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells ( intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into many compartments. In the eukaryotic cell, the cytosol is surrounded by the cell membrane and is part of the cytoplasm, which also comprises the mitochondria, plastids, and other organelles (but not their internal fluids and structures); the cell nucleus is separate. The cytosol is thus a liquid matrix around the organelles. In prokaryotes, most of the chemical reactions of metabolism take place in the cytosol, while a few take place in membranes or in the periplasmic space. In eukaryotes, while many metabolic pathways still occur in the cytosol, others take place within organelles. The cytosol is a complex mixture of substances dissolved in water. Although water forms the large majority of the cytosol, its structure and proper ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
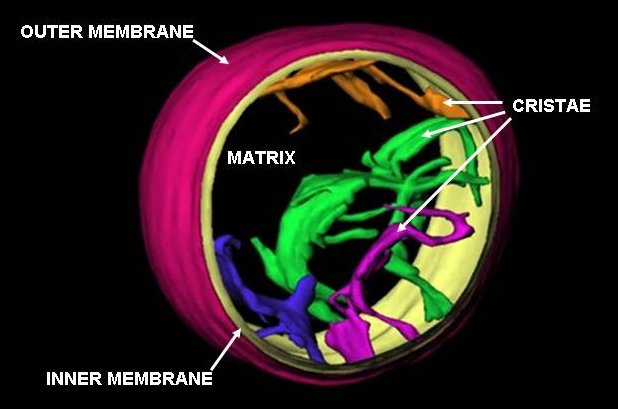
Mitochondria
A mitochondrion () is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'', meaning a thread-like granule, was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase popularized by Philip Siekevitz in a 1957 ''Scientific American'' article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). The multicellular animal '' Henneguya salminicola'' is known to have retained mitochondrion-related organelles despite a complete loss of their mitochondrial genome. A large number of unicellular organisms, such as microspo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Urea Cycle
The urea cycle (also known as the ornithine cycle) is a cycle of biochemical reactions that produces urea (NH2)2CO from ammonia (NH3). Animals that use this cycle, mainly amphibians and mammals, are called ureotelic. The urea cycle converts highly toxic ammonia to urea for excretion. This cycle was the first metabolic cycle to be discovered by Hans Krebs and Kurt Henseleit in 1932, five years before the discovery of the TCA cycle. The urea cycle was described in more detail later on by Ratner and Cohen. The urea cycle takes place primarily in the liver and, to a lesser extent, in the kidneys. Function Amino acid catabolism results in waste ammonia. All animals need a way to excrete this product. Most aquatic organisms, or ammonotelic organisms, excrete ammonia without converting it. Organisms that cannot easily and safely remove nitrogen as ammonia convert it to a less toxic substance, such as urea, via the urea cycle, which occurs mainly in the liver. Urea produced by the li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ornithine Translocase Deficiency
Ornithine translocase deficiency, also called hyperornithinemia-hyperammonemia-homocitrullinuria (HHH) syndrome, is a rare autosomal recessive urea cycle disorder affecting the enzyme ornithine translocase, which causes ammonia to accumulate in the blood, a condition called hyperammonemia. Ammonia, which is formed when proteins are broken down in the body, is toxic if the levels become too high. The nervous system is especially sensitive to the effects of excess ammonia. Signs and symptoms Pathophysiology Mutations in '' SLC25A15'' cause ornithine translocase deficiency. Ornithine translocase deficiency belongs to a class of metabolic disorders referred to as urea cycle disorders. The urea cycle is a sequence of reactions that occurs in liver cells. This cycle processes excess nitrogen, generated when protein is used by the body, to make a compound called urea that is excreted by the kidneys. The ''SLC25A15'' gene provides instructions for making a protein called a mitocho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hyperammonemia
Hyperammonemia, or high ammonia levels, is a metabolic disturbance characterised by an excess of ammonia in the blood. Severe hyperammonemia is a dangerous condition that may lead to brain injury and death. It may be primary or secondary. Ammonia is a substance that contains nitrogen. It is a product of the catabolism of protein. It is converted to the less toxic substance urea prior to excretion in urine by the kidneys. The metabolic pathways that synthesize urea involve reactions that start in the mitochondria and then move into the cytosol. The process is known as the urea cycle, which comprises several enzymes acting in sequence. It is greatly exacerbated by common zinc deficiency, which raises ammonia levels further. Levels Normal blood ammonia levels in adults range from 20 to 50μmol/L or less than 26 to 30μmol/L. There is at present no clear scientific consensus on the upper limits of ammonia levels for different age groups. In any case, hyperammonemia is generally defi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translocase
Translocase is a general term for a protein that assists in moving another molecule, usually across a cell membrane. These Enzyme, enzymes catalyze the movement of ions or molecules across membranes or their separation within membranes. The reaction is designated as a transfer from “side 1” to “side 2” because the designations “in” and “out”, which had previously been used, can be ambiguous. Translocases are the most common Bacterial secretion system, secretion system in Gram positive bacteria. It is also a historical term for the protein now called elongation factor G, due to its function in moving the transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome. History The enzyme classification and nomenclature list was first approved by the International Union of Biochemistry in 1961. Six enzyme classes had been recognized based on the type of chemical reaction catalyzed, including Oxidoreductase, oxidoreductases (EC 1), Transferase, transferases (EC 2), ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Solute Carrier Family
The solute carrier (SLC) group of membrane transport proteins include over 400 members organized into 66 families. Most members of the SLC group are located in the cell membrane. The SLC gene nomenclature system was originally proposed by the HUGO Gene Nomenclature Committee (HGNC) and is the basis for the official HGNC names of the genes that encode these transporters. A more general transmembrane transporter classification can be found in TCDB, TCDB database. Solutes that are transported by the various SLC group members are extremely diverse and include both charged and uncharged organic molecules as well as inorganic ions and the gas Ammonia transporter, ammonia. As is typical of integral membrane proteins, SLCs contain a number of hydrophobic transmembrane Alpha helix, alpha helices connected to each other by hydrophilic intra- and extra-cellular loops. Depending on the SLC, these transporters are functional as either monomers or obligate homo- or hetero-oligomers. Many SLC fam ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |