|
Non-proteinogenic Amino Acids
In biochemistry, non-coded or non-proteinogenic amino acids are distinct from the 22 proteinogenic amino acids (21 in eukaryotesplus formylmethionine in eukaryotes with prokaryote organelles like mitochondria), which are naturally encoded in the genome of organisms for the assembly of proteins. However, over 140 non-proteinogenic amino acids occur naturally in proteins and thousands more may occur in nature or be synthesized in the laboratory. Chemically synthesized amino acids can be called unnatural amino acids. Unnatural amino acids can be synthetically prepared from their native analogs via modifications such as amine alkylation, side chain substitution, structural bond extension cyclization, and isosteric replacements within the amino acid backbone. Many non-proteinogenic amino acids are important: * intermediates in biosynthesis, * in post-translational formation of proteins, * in a physiological role (e.g. components of Peptidoglycan, bacterial cell walls, neurotransmitters ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nonproteinogenic AAs
Proteinogenic amino acids are amino acids that are incorporated biosynthetically into proteins during translation (biology), translation from RNA. The word "proteinogenic" means "protein creating". Throughout known life, there are 22 genetically encoded (proteinogenic) amino acids, 20 in the standard genetic code and an additional 2 (selenocysteine and pyrrolysine) that can be incorporated by special translation mechanisms. In contrast, non-proteinogenic amino acids are amino acids that are either not incorporated into proteins (like Gamma-aminobutyric acid, GABA, L-DOPA, L-DOPA, or triiodothyronine), misincorporated in place of a genetically encoded amino acid, or not produced directly and in isolation by standard cellular machinery (like hydroxyproline). The latter often results from post-translational modification of proteins. Some non-proteinogenic amino acids are incorporated into nonribosomal peptides which are synthesized by non-ribosomal peptide synthetases. Both eukaryote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stereoisomerism
In stereochemistry, stereoisomerism, or spatial isomerism, is a form of isomerism in which molecules have the same molecular formula and sequence of bonded atoms (constitution), but differ in the three-dimensional orientations of their atoms in space. This contrasts with structural isomers, which share the same molecular formula, but the bond connections or their order differs. By definition, molecules that are stereoisomers of each other represent the same structural isomer. Enantiomers Enantiomers, also known as optical isomers, are two stereoisomers that are related to each other by a reflection: they are mirror images of each other that are non-superposable. Human hands are a macroscopic analog of this. Every stereogenic center in one has the opposite configuration in the other. Two compounds that are enantiomers of each other have the same physical properties, except for the direction in which they rotate polarized light and how they interact with different enantiomers of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
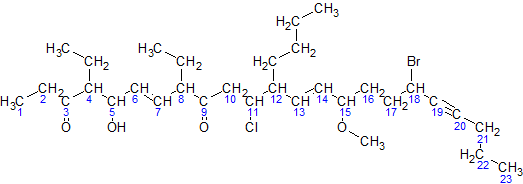
IUPAC Nomenclature Of Organic Chemistry
In chemical nomenclature, the IUPAC nomenclature of organic chemistry is a method of naming organic chemical compounds as recommended by the International Union of Pure and Applied Chemistry (IUPAC). It is published in the '' Nomenclature of Organic Chemistry'' (informally called thBlue Book. Ideally, every possible organic compound should have a name from which an unambiguous structural formula can be created. There is also an IUPAC nomenclature of inorganic chemistry. To avoid long and tedious names in normal communication, the official IUPAC naming recommendations are not always followed in practice, except when it is necessary to give an unambiguous and absolute definition to a compound. IUPAC names can sometimes be simpler than older names, as with ethanol, instead of ethyl alcohol. For relatively simple molecules they can be more easily understood than non-systematic names, which must be learnt or looked over. However, the common or trivial name is often substantially ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Post-translational Modification
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translation (biology), translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signal transduction, signalling, as for example when prohormones are converted to hormones. Post-translational modifications can occur on the amino acid side chains or at the protein's C-terminus, C- or N-terminus, N- termini. They can expand the chemical set of the 22 proteinogenic amino acid, amino acids by changing an existing functional group or adding a new one such as phosphate. Phosphorylation is highly effective for controlling the enzyme activity and is the most common change after translation. Many eukaryotic and prokaryotic proteins also have carbohydrate molecules attached to them in a pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ornithine
Ornithine is a non-proteinogenic α-amino acid that plays a role in the urea cycle. It is not incorporated into proteins during translation. Ornithine is abnormally accumulated in the body in ornithine transcarbamylase deficiency, a disorder of the urea cycle. The Moiety (chemistry), moiety derived from ornithine is called ornithyl. Role in urea cycle L-Ornithine is one of the products of the action of the enzyme arginase on L-arginine, creating urea. Therefore, ornithine is a central component of the urea cycle, which enables the disposal of excess nitrogen. Ornithine itself is recycled and, in a sense, acts as a catalyst. First, ammonia is converted into carbamoyl phosphate () by carbamoyl phosphate synthetase. Ornithine transcarbamylase then catalyzes the reaction between carbamoyl phosphate and ornithine to form citrulline and phosphate (Pi). Another amino group is contributed by aspartate, leading to the formation of arginine and the byproduct fumarate. The resulting arginine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Norleucine
Norleucine (abbreviated as Nle) is an amino acid with the formula CH3(CH2)3CH(NH2)CO2H. A systematic name for this compound is 2-aminohexanoic acid. The compound is an isomer of the more common amino acid leucine. Like most other α-amino acids, norleucine is chiral. It is a white, water-soluble solid. Occurrence Together with norvaline, norleucine is found in small amounts in some bacterial strains where its concentration can approach millimolar. Its biosynthesis has been examined. It arises via the action of 2-isopropylmalate synthase on α-ketobutyrate. The incorporation of Nle into peptides reflects the imperfect selectivity of the associated aminoacyl-tRNA synthetase. In Miller–Urey experiments probing prebiotic synthesis of amino acids, norleucine and especially norvaline are formed. Uses It is nearly isosteric with methionine, even though it does not contain sulfur. For this reason, norleucine has been used to probe the role of methionine in Beta amyloid, Amyloid-β p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PYLIS Downstream Sequence
In biology, the PYLIS downstream sequence (PYLIS: ''pyrrolysine insertion sequence'') is a stem-loop structure that appears on some mRNA sequences. This structural motif was previously thought to cause the UAG (amber) stop codon to be translated to the amino acid pyrrolysine instead of ending the protein translation. However, it has been shown that PYLIS has no effect upon the efficiency of the UAG suppression, hence even its name is, in fact, incorrect. See also *SECIS element In biology, the SECIS element (SECIS: ''selenocysteine insertion sequence'') is an RNA element around 60 nucleotides in length that adopts a stem-loop structure. This structural motif (pattern of nucleotides) directs the cell to translate ... References Further reading * * External links * Cis-regulatory RNA elements {{Genetics-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SECIS Element
In biology, the SECIS element (SECIS: ''selenocysteine insertion sequence'') is an RNA element around 60 nucleotides in length that adopts a stem-loop structure. This structural motif (pattern of nucleotides) directs the cell to translate UGA codons as selenocysteines (UGA is normally a stop codon). SECIS elements are thus a fundamental aspect of messenger RNAs encoding selenoproteins, proteins that include one or more selenocysteine residues. Location and function In bacteria the SECIS element appears soon after the UGA codon it affects. In archaea and eukaryotes, it occurs in the 3' UTR of an mRNA, and can cause multiple UGA codons within the mRNA to code for selenocysteine. One archaeal SECIS element, in '' Methanococcus,'' is located in the 5' UTR. In any case, it serves to recruit EEFSEC or SelB, the specialized homolog of EF-Tu/eEF1&alpha, with the ability to read tRNASec. Characteristics The SECIS elements appear defined by sequence characteristics (parti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrrolysine
Pyrrolysine (symbol Pyl or O), encoded by the 'amber' stop codon UAG, is a proteinogenic amino acid that is used in some methanogenic archaea and in bacteria. It consists of lysine with a 4-methylpyrroline-5-carboxylate in amide linkage with the εN of the lysine. Its pyrroline side-chain is similar to that of lysine in being basic and positively charged at neutral pH. Genetics Nearly all genes are translated using only 20 standard amino acid building blocks. Two unusual genetically-encoded amino acids are selenocysteine and pyrrolysine. Pyrrolysine was discovered in 2002 at the active site of methyltransferase enzyme from a methane-producing archeon, '' Methanosarcina barkeri''. This amino acid is encoded by UAG (normally a stop codon), and its synthesis and incorporation into protein is mediated via the biological machinery encoded by the ''pylTSBCD'' cluster of genes. Synthesis Pyrrolysine is synthesized ''in vivo'' by joining two molecules of L-lysine. One molecule of ly ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Selenocysteine
Selenocysteine (symbol Sec or U, in older publications also as Se-Cys) is the 21st proteinogenic amino acid. Selenoproteins contain selenocysteine residues. Selenocysteine is an analogue of the more common cysteine with selenium in place of the sulfur. Selenocysteine is present in several enzymes (for example glutathione peroxidases, tetraiodothyronine 5 deiodinase, tetraiodothyronine 5′ deiodinases, thioredoxin reductases, formate dehydrogenases, glycine reductases, selenophosphate synthetase 2, methionine-''R''-sulfoxide reductase B1 (SEPX1), and some hydrogenases). It occurs in all three Domain (biology), domains of life, including important enzymes (listed above) present in humans. Selenocysteine was discovered in 1974 by biochemist Thressa Stadtman at the National Institutes of Health. Chemistry Selenocysteine is the Se-analogue of cysteine. It is rarely encountered outside of living tissue (nor is it available commercially) because of its high susceptiblility to air-oxi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (biology)
In biology, translation is the process in living Cell (biology), cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotides in the RNA. The nucleotides are considered three at a time. Each such triple results in the addition of one specific amino acid to the protein being generated. The matching from nucleotide triple to amino acid is called the genetic code. The translation is performed by a large complex of functional RNA and proteins called ribosomes. The entire process is called gene expression. In translation, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later protein folding, folds into an Activation energy, active protein and performs its functions in the cell. The polypeptide can also start folding during protein synthesis. The ribosome facilitates decoding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Imino Acid
In organic chemistry, an imino acid is any molecule that contains both imine (>C=NH) and carboxyl (-C(=O)-OH) functional groups. Imino acids are structurally related to amino acids, which have amino group instead of imine—a difference of single vs double-bond between nitrogen and carbon. The simplest example is dehydroglycine. D-Amino acid oxidase is an enzyme that is able to convert amino acids into imino acids. Also the direct biosynthetic precursor to the amino acid proline is the imino acid (''S'')-Δ1-pyrroline-5-carboxylate (P5C). Related terminology Secondary amino acids, amino acids containing a secondary amine group are sometimes named imino acids, though this usage is obsolescent. The only proteinogenic amino acid of this type is proline, although the related non-proteinogenic amino acids hydroxyproline (2''S'',4''R'')-4-Hydroxyproline, or L-hydroxyproline ( C5 H9 O3 N), is an amino acid, abbreviated as Hyp or O, ''e.g.'', in Protein Data Bank. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |