|
NEDD8 Activating Enzyme
NEDD8 is a protein that in humans is encoded by the ''NEDD8'' gene. (in ''saccharomyces cerevisiae'' this protein is known as Rub1) This ubiquitin-like (UBL) protein becomes covalently conjugated to a limited number of cellular proteins, in a process called NEDDylation similar to ubiquitination. Human NEDD8 shares 60% amino acid sequence identity to ubiquitin. The primary known substrates of NEDD8 modification are the cullin subunits of cullin-based E3 ubiquitin ligases, which are active only when NEDDylated. Their NEDDylation is critical for the recruitment of E2 to the ligase complex, thus facilitating ubiquitin conjugation. NEDD8 modification has therefore been implicated in cell cycle progression and cytoskeletal regulation. Activation and conjugation As with ubiquitin and SUMO, NEDD8 is conjugated to cellular proteins after its C-terminal tail is processed. The NEDD8 activating E1 enzyme is a heterodimer composed of APPBP1 and UBA3 subunits. The APPBP1/UBA3 enzyme has homolog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid resid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DDB2
DNA damage-binding protein 2 is a protein that in humans is encoded by the ''DDB2'' gene. Structure As indicated by Rapić-Otrin et al. in 2003, the ''DDB2'' gene is located on human chromosome 11p11.2, spans a region of approximately 24 – 26 kb and includes 10 exons. The DDB2 protein contains five putative WD40 repeats (sequences of about 40 amino acids that can interact with each other) positioned downstream from the second exon. The WD40 motif identified in DDB2 is characteristic of proteins involved in the recognition of chromatin proteins. The C-terminal region of DDB2 (a 48 kDa molecular weight protein) is essential for binding to DDB1 (a larger 127 kDa protein). Together, the two proteins form a UV-damaged DNA binding protein complex (UV-DDB). Deficiency in humans If humans have a mutation in each copy of their ''DDB2'' gene, this causes a mild form of the human disease xeroderma pigmentosum, called XPE. Patients in the XPE group have mild dermatological manifestatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adipogenesis
Adipogenesis is the formation of adipocytes (fat cells) from stem cells. It involves 2 phases, determination, and terminal differentiation. Determination is mesenchymal stem cells committing to the adipocyte precursor cells, also known as preadipocytes which lose the potential to differentiate to other types of cells such as chondrocytes, myocytes, and osteoblasts. Terminal differentiation is that preadipocytes differentiate into mature adipocytes. Adipocytes can arise either from preadipocytes resident in adipose tissue, or from bone-marrow derived progenitor cells that migrate to adipose tissue. Introduction Adipocytes play a vital role in energy homeostasis and process the largest energy reserve as triglycerol in the body of animals. Adipocytes stay in a dynamic state, they start expanding when the energy intake is higher than the expenditure and undergo mobilization when the energy expenditure exceeds the intake. This process is highly regulated by counter regulatory hormo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peroxisome Proliferator-activated Receptor Gamma
Peroxisome proliferator- activated receptor gamma (PPAR-γ or PPARG), also known as the glitazone reverse insulin resistance receptor, or NR1C3 (nuclear receptor subfamily 1, group C, member 3) is a type II nuclear receptor functioning as a transcription factor that in humans is encoded by the ''PPARG'' gene. Tissue distribution PPARG is mainly present in adipose tissue, colon and macrophages. Two isoforms of PPARG are detected in the human and in the mouse: PPAR-γ1 (found in nearly all tissues except muscle) and PPAR-γ2 (mostly found in adipose tissue and the intestine). Gene expression This gene encodes a member of the peroxisome proliferator-activated receptor (PPAR) subfamily of nuclear receptors. PPARs form heterodimers with retinoid X receptors (RXRs) and these heterodimers regulate transcription of various genes. Three subtypes of PPARs are known: PPAR-alpha, PPAR-delta, and PPAR-gamma. The protein encoded by this gene is PPAR-gamma and is a regulator of adipocyte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phases Of Clinical Research
The phases of clinical research are the stages in which scientists conduct experiments with a health intervention to obtain sufficient evidence for a process considered effective as a medical treatment. For drug development, the clinical phases start with testing for safety in a few human subjects, then expand to many study participants (potentially tens of thousands) to determine if the treatment is effective. Clinical research is conducted on drug candidates, vaccine candidates, new medical devices, and new diagnostic assays. Summary Clinical trials testing potential medical products are commonly classified into four phases. The drug development process will normally proceed through all four phases over many years. If the drug successfully passes through Phases I, II, and III, it will usually be approved by the national regulatory authority for use in the general population. Phase IV trials are 'post-marketing' or 'surveillance' studies conducted to monitor safety over sever ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pevonedistat
Pevonedistat (MLN4924) is a selective NEDD8 inhibitor. It is being investigated as a cancer treatment, e.g. for mantle cell lymphoma (MCL). Target of pevonedistat NEDD8-activating enzyme (NAE) is a heterodimeric molecule consisting of amyloid beta precursor protein-binding protein 1 (APPBP1) and ubiquitin-like modifier activating enzyme 3 ( UBA3). Material was copied from this source, which is available under Creative Commons Attribution 4.0 International (CC BY 4.0)license. As reviewed by Xu et al., in a first step NAE binds ATP and NEDD8 and catalyzes the formation of a NEDD8-AMP intermediate. This intermediate binds the adenylation domain of NAE. NEDD8-AMP reacts with the catalytic cysteine in UBA3 during which NEDD8 is transferred to the catalytic cysteine, resulting in a high energy thioester linkage. NAE then binds ATP and NEDD8 to generate a second NEDD8-AMP, forming a fully loaded NAE carrying two activated NEDD8 molecules (i.e., one as a thioester and the other as a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
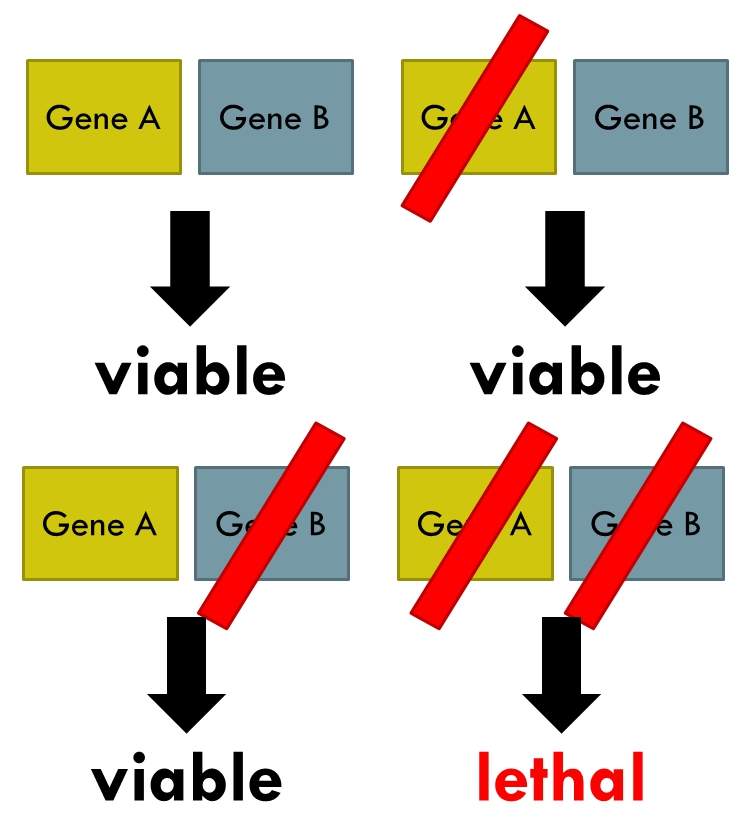
Synthetic Lethality
Synthetic lethality is defined as a type of genetic interaction where the combination of two genetic events results in cell death or death of an organism. Although the foregoing explanation is wider than this, it is common when referring to synthetic lethality to mean the situation arising by virtue of a combination of deficiencies of two or more genes leading to cell death (whether by means of apoptosis or otherwise), whereas a deficiency of only one of these genes does not. In a synthetic lethal genetic screen, it is necessary to begin with a mutation that does not result in cell death, although the effect of that mutation could result in a differing phenotype (slow growth for example), and then systematically test other mutations at additional loci to determine which, in combination with the first mutation, causes cell death arising by way of deficiency or abolition of expression. Synthetic lethality has utility for purposes of molecular targeted cancer therapy. The first example ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-homologous End Joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair(HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber. NHEJ is typically guided by short homologous DNA sequences called microhomologies. These microhomologies are often present in single-stranded overhangs on the ends of double-strand breaks. When the overhangs are perfectly compatible, NHEJ usually repairs the break accurately. Imprecise repair leading to loss of nucleotides can also occur, but is much more common when the overhangs are not compatible. Inappropriate NHEJ can lead to translocations and telomere fusion, hallmarks ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Methylation In Cancer
DNA methylation in cancer plays a variety of roles, helping to change the healthy cells by regulation of gene expression to a cancer cells or a diseased cells Regulation of transcription in cancer, disease pattern. One of the most widely studied DNA methylation dysregulation is the promoter hypermethylation where the CPGs islands in the promoter regions are methylated contributing or causing genes to be silenced. All mammalian cells descended from a fertilized egg (a zygote) share a common DNA sequence (except for new mutations in some lineages). However, during development and formation of different tissues Epigenetics, epigenetic factors change. The changes include histone modifications, CpG site#CpG island, CpG island methylations and chromatin reorganizations which can cause the stable silencing or activation of particular genes. Once differentiated tissues are formed, CpG island methylation is generally stably inherited from one cell division to the next through the DNA methyl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cancer Epigenetics
Cancer epigenetics is the study of epigenetic modifications to the DNA of cancer cells that do not involve a change in the nucleotide sequence, but instead involve a change in the way the genetic code is expressed. Epigenetic mechanisms are necessary to maintain normal sequences of tissue specific gene expression and are crucial for normal development. They may be just as important, if not even more important, than genetic mutations in a cell's transformation to cancer. The disturbance of epigenetic processes in cancers, can lead to a loss of expression of genes that occurs about 10 times more frequently by transcription silencing (caused by epigenetic promoter hypermethylation of CpG islands) than by mutations. As Vogelstein et al. points out, in a colorectal cancer there are usually about 3 to 6 driver mutations and 33 to 66 hitchhiker or passenger mutations. However, in colon tumors compared to adjacent normal-appearing colonic mucosa, there are about 600 to 800 heavily methyl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Instability
Genome instability (also genetic instability or genomic instability) refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy. The sources of genome instability have only recently begun to be elucidated. A high frequency of externally caused DNA damage can be one source of genome instability since DNA damage can cause inaccurate translesion DNA synthesis past the damage or errors in repair, leading to mutation. Another source of genome instability may be epigenetic or mutational reductions in expression of DNA repair genes. Because endogenous (metabolically-caused) DNA damage is very fr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulation Of Transcription In Cancer
Generally, in progression to cancer, hundreds of genes are silenced or activated. Although silencing of some genes in cancers occurs by mutation, a large proportion of carcinogenic gene silencing is a result of altered DNA methylation (see DNA methylation in cancer). DNA methylation causing silencing in cancer typically occurs at multiple CpG sites in the CpG islands that are present in the promoters of protein coding genes. Altered expressions of microRNAs also silence or activate many genes in progression to cancer (see microRNAs in cancer). Altered microRNA expression occurs through hyper/hypo-methylation of CpG sites in CpG islands in promoters controlling transcription of the microRNAs. Silencing of DNA repair genes through methylation of CpG islands in their promoters appears to be especially important in progression to cancer (see methylation of DNA repair genes in cancer). CpG islands in promoters In humans, about 70% of promoters located near the transcription sta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |