|
Mir-29 MicroRNA Precursor
The miR-29 microRNA precursor, or pre-miRNA, is a small RNA molecule in the shape of a stem-loop or Hairpin (fashion), hairpin. Each arm of the hairpin can be processed into one member of a closely related family of short non-coding RNAs that are involved in regulating gene expression. The processed, or "mature" products of the precursor molecule are known as microRNA (miRNA), and have been predicted or confirmed in a wide range of speciessee 'MIPF0000009' in miRBase: the microRNA database. miRNA processing Animal miRNAs are first transcribed as a primary miRNA molecule. This "pri-miRNA" may contain one or more precursor hairpins, which are freed from the pri-miRNA by the nuclear enzyme Drosha. The approximately 70 nucleotide precursor hairpin is exported from the nucleus and subsequently processed by the Dicer enzyme to give a mature miRNA that is on average 22 nucleotides long. Either arm of the precursor may yield a mature RNA, although either the 3' (3p) or the 5' (5p) arm is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MCL1
Induced myeloid leukemia cell differentiation protein Mcl-1 is a protein that in humans is encoded by the ''MCL1'' gene. Function The protein encoded by this gene belongs to the Bcl-2 family. Alternative splicing occurs at this locus and two transcript variants encoding distinct isoforms have been identified. The longer gene product (isoform 1) enhances cell survival by inhibiting apoptosis while the alternatively spliced shorter gene product (isoform 2) promotes apoptosis and is death-inducing. The protein MCL1 has a very short biological half-life of only 20–30 minutes. The loss of MCL1 has a more dramatic impact than the loss of any other anti-apoptotic member of the Bcl-2 family. Loss of the ''Mcl-1'' gene results in embryo death when the embryo is only around 3.5 days old, before it has even implanted. Conditional deletion of ''Mcl-1'' depletes a wide variety of cells, including hematopoietic stem cells, B cell–committed progenitors, T cell–committed progenitors, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Voltage-dependent Anion Channel
Voltage-dependent anion channels, or mitochondrial porins, are a class of porin ion channel located on the outer mitochondrial membrane. There is debate as to whether or not this channel is expressed in the cell surface membrane. This major protein of the outer mitochondrial membrane of eukaryotes forms a voltage-dependent anion-selective channel (VDAC) that behaves as a general diffusion pore for small hydrophilic molecules. The channel adopts an open conformation at low or zero membrane potential and a closed conformation at potentials above 30–40 mV. VDAC facilitates the exchange of ions and molecules between mitochondria and cytosol and is regulated by the interactions with other proteins and small molecules. Structure This protein contains about 280 amino acids and forms a beta barrel which spans the mitochondrial outer membrane. Since its discovery in 1976, extensive function and structure analysis of VDAC proteins has been conducted. A prominent feature of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stable Isotope Labeling By Amino Acids In Cell Culture
Stable isotope labeling by/with amino acids in cell culture (SILAC) is a technique based on mass spectrometry that detects differences in protein abundance among samples using non-radioactive isotopic labeling. It is a popular method for quantitative proteomics. Procedure Two populations of cells are cultivated in cell culture. One of the cell populations is fed with growth medium containing normal amino acids. In contrast, the second population is fed with growth medium containing amino acids labeled with stable (non-radioactive) heavy isotopes. For example, the medium can contain arginine labeled with six carbon-13 atoms (13C) instead of the normal carbon-12 (12C). When the cells are growing in this medium, they incorporate the heavy arginine into all of their proteins. Thereafter, all peptides containing a single arginine are 6 Da heavier than their normal counterparts. Alternatively, uniform labeling with 13C or 15N can be used. Proteins from both cell populations are combined ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quantitative Proteomics
Quantitative proteomics is an analytical chemistry technique for determining the amount of proteins in a sample. The methods for protein identification are identical to those used in general (i.e. qualitative) proteomics, but include quantification as an additional dimension. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about the physiological differences between two biological samples. For example, this approach can be used to compare samples from healthy and diseased patients. Quantitative proteomics is mainly performed by two-dimensional gel electrophoresis (2-DE), preparative native PAGE, or mass spectrometry (MS). However, a recent developed method of quantitative dot blot (QDB) analysis is able to measure both the absolute and relative quantity of an individual proteins in the sample in high throughput format, thus open a new direction for proteomic research. In contrast to 2-DE, which requires MS f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ZFP36
Tristetraprolin (TTP), also known as zinc finger protein 36 homolog (ZFP36), is a protein that in humans, mice and rats is encoded by the ''ZFP36'' gene. It is a member of the TIS11 ( TPA-induced sequence) family, along with butyrate response factors 1 and 2. TTP binds to AU-rich elements (AREs) in the 3'-untranslated regions (UTRs) of the mRNAs of some cytokines and promotes their degradation. For example, TTP is a component of a negative feedback loop that interferes with TNF-alpha production by destabilizing its mRNA. Mice deficient in TTP develop a complex syndrome of inflammatory diseases. Interactions ZFP36 has been shown to interact with 14-3-3 protein family members, such as YWHAH, and with NUP214, a member of the nuclear pore complex. Regulation Post-transcriptionally, TTP is regulated in several ways. The subcellular localization of TTP is influenced by interactions with protein partners such as the 14-3-3 family of proteins. These interactions and, possibly, inter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lung Cancer
Lung cancer, also known as lung carcinoma, is a malignant tumor that begins in the lung. Lung cancer is caused by genetic damage to the DNA of cells in the airways, often caused by cigarette smoking or inhaling damaging chemicals. Damaged airway cells gain the ability to multiply unchecked, causing the growth of a tumor. Without treatment, tumors spread throughout the lung, damaging lung function. Eventually lung tumors metastasize, spreading to other parts of the body. Early lung cancer often has no symptoms and can only be detected by medical imaging. As the cancer progresses, most people experience nonspecific respiratory problems: coughing, shortness of breath, or chest pain. Other symptoms depend on the location and size of the tumor. Those suspected of having lung cancer typically undergo a series of imaging tests to determine the location and extent of any tumors. Definitive diagnosis of lung cancer requires a biopsy of the suspected tumor be examined by a patholo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNMT3B
DNA (cytosine-5)-methyltransferase 3 beta, is an enzyme that in humans in encoded by the DNMT3B gene. Mutation in this gene are associated with immunodeficiency, centromere instability and facial anomalies syndrome. Function CpG methylation is an epigenetic modification that is important for embryonic development, imprinting, and X-chromosome inactivation. Studies in mice have demonstrated that DNA methylation is required for mammalian development. This gene encodes a DNA methyltransferase which is thought to function in ''de novo'' methylation, rather than maintenance methylation. The protein localizes primarily to the nucleus and its expression is developmentally regulated. Eight alternatively spliced transcript variants have been described. The full length sequences of variants 4 and 5 have not been determined. Clinical significance Immunodeficiency-centromeric instability-facial anomalies (ICF) syndrome is a result of defects in lymphocyte maturation resulting from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNMT3A
DNA (cytosine-5)-methyltransferase 3A (DNMT3A) is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. The enzyme is encoded in humans by the ''DNMT3A'' gene. This enzyme is responsible for ''wikt:de novo, de novo'' DNA methylation. Such function is to be distinguished from maintenance DNA methylation which ensures the fidelity of replication of inherited epigenetic patterns. DNMT3A forms part of the family of DNA methyltransferase enzymes, which consists of the protagonists DNMT1, DNMT3A and DNMT3B. While ''de novo'' DNA methylation modifies the information passed on by the parent to the progeny, it enables key Epigenetics, epigenetic modifications essential for processes such as cellular differentiation and Embryogenesis, embryonic development, transcriptional regulation, heterochromatin formation, X-inactivation, Genomic imprinting, imprinting and genome stability. ''DNMT3a'' is the gene most commonly fou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor. Classification Substrate MTases can be divided into three different groups on the basis of the chemical reactions they catalyze: * m6A - those that generate N6-methyladenine * m4C - those that generate N4-methylcytosine * m5C - those that generate C5-methylcytosine m6A and m4C methyltransferases are found primarily in prokaryotes (although recent evidence has suggested that m6A is abundant in eukaryotes). m5C methyltransferases are found in some lower eukaryotes, in most higher plants, and in animals beginning with the echinoderms. The m6A methyltransferases (N-6 adenine-specific DNA methylase) (A-Mtase) are enzymes that specifically methylate the amino group at the C-6 position of adenine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leukemias
Leukemia ( also spelled leukaemia; pronounced ) is a group of blood cancers that usually begin in the bone marrow and produce high numbers of abnormal blood cells. These blood cells are not fully developed and are called ''blasts'' or '' leukemia cells''. Symptoms may include bleeding and bruising, bone pain, fatigue, fever, and an increased risk of infections. These symptoms occur due to a lack of normal blood cells. Diagnosis is typically made by blood tests or bone marrow biopsy. The exact cause of leukemia is unknown. A combination of genetic factors and environmental (non-inherited) factors are believed to play a role. Risk factors include smoking, ionizing radiation, petrochemicals (such as benzene), prior chemotherapy, and Down syndrome. People with a family history of leukemia are also at higher risk. There are four main types of leukemia—acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic lymphocytic leukemia (CLL) and chronic myeloid leuk ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
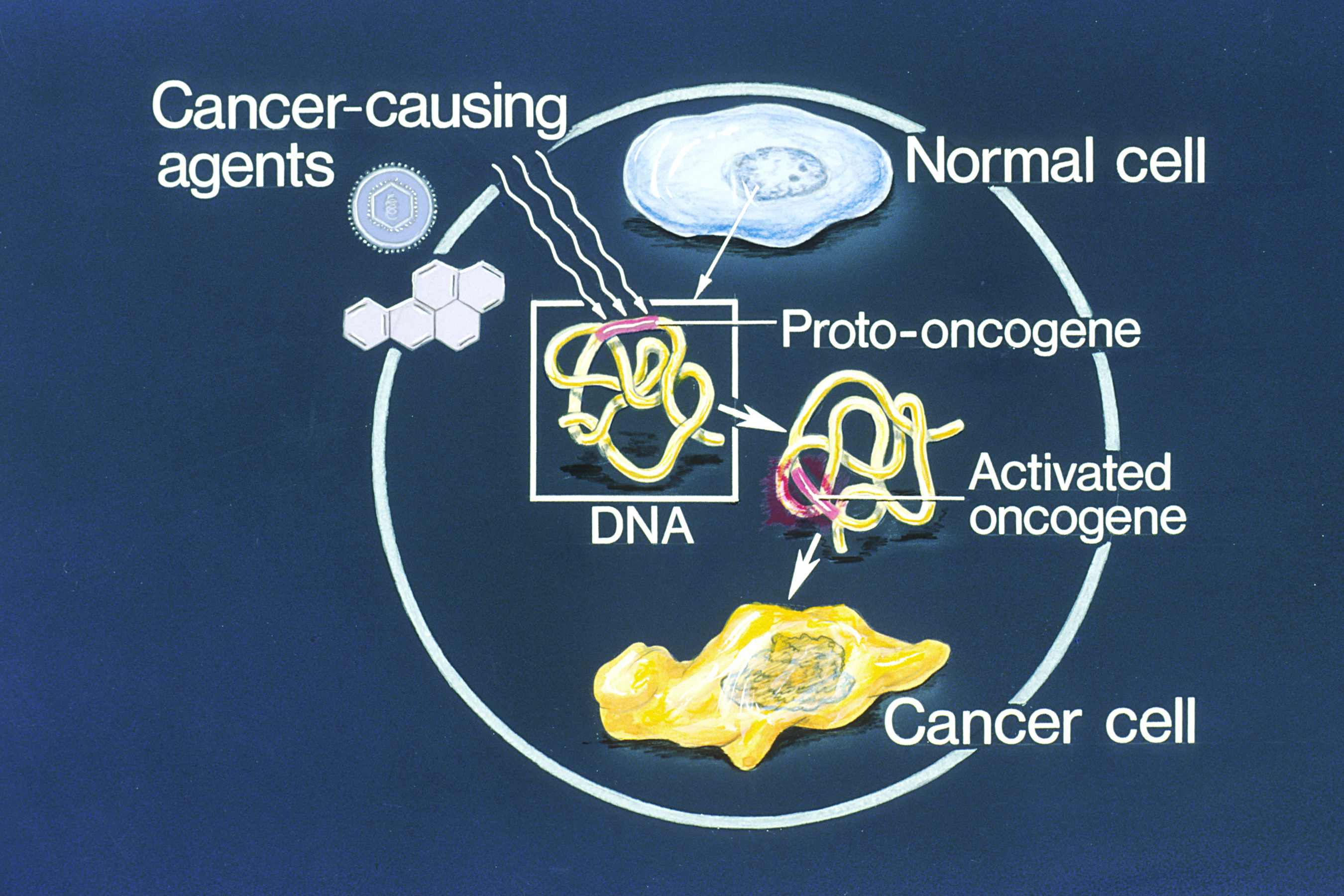
Oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.Kimball's Biology Pages. "Oncogenes" Free full text Most normal cells undergo a preprogrammed rapid cell death () if critical functions are altered and then malfunction. Activated oncogenes can cause those cells designated for apoptosis to survive and proliferate instead. Most oncogenes began as proto-oncogenes: normal genes involved in cell growth and proliferation or inhibition of apoptosis. If, through mutation, normal genes promoting cellular growth are up-regulated (gain-of-function mutation), they predispose the cel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |