 |
Micro-arrays For Mass Spectrometry
In cell biology, single-cell analysis and subcellular analysis refer to the study of genomics, transcriptomics, proteomics, metabolomics, and cell–cell interactions at the level of an individual cell, as opposed to more conventional methods which study bulk populations of many cells. The concept of single-cell analysis originated in the 1970s. Before the discovery of heterogeneity, single-cell analysis mainly referred to the analysis or manipulation of an individual cell within a bulk population of cells under the influence of a particular condition using optical or electron microscopy. Due to the heterogeneity seen in both eukaryotic and prokaryotic cell populations, analyzing the biochemical processes and features of a single cell makes it possible to discover mechanisms which are too subtle or infrequent to be detectable when studying a bulk population of cells; in conventional multi-cell analysis, this variability is usually masked by the average behavior of the large ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
 |
Proteinsynthesis
Protein biosynthesis, or protein synthesis, is a core biological process, occurring inside cells, balancing the loss of cellular proteins (via degradation or export) through the production of new proteins. Proteins perform a number of critical functions as enzymes, structural proteins or hormones. Protein synthesis is a very similar process for both prokaryotes and eukaryotes but there are some distinct differences. Protein synthesis can be divided broadly into two phases: transcription and translation. During transcription, a section of DNA encoding a protein, known as a gene, is converted into a molecule called messenger RNA (mRNA). This conversion is carried out by enzymes, known as RNA polymerases, in the nucleus of the cell. In eukaryotes, this mRNA is initially produced in a premature form (pre-mRNA) which undergoes post-transcriptional modifications to produce mature mRNA. The mature mRNA is exported from the cell nucleus via nuclear pores to the cytoplasm of the cell fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
 |
Raman Spectroscopy
Raman spectroscopy () (named after physicist C. V. Raman) is a Spectroscopy, spectroscopic technique typically used to determine vibrational modes of molecules, although rotational and other low-frequency modes of systems may also be observed. Raman spectroscopy is commonly used in chemistry to provide a structural fingerprint by which molecules can be identified. Raman spectroscopy relies upon inelastic scattering of photons, known as Raman scattering. A source of monochromatic light, usually from a laser in the visible spectrum, visible, near infrared, or ultraviolet, near ultraviolet range is used, although X-ray Raman scattering, X-rays can also be used. The laser light interacts with molecular vibrations, phonons or other excitations in the system, resulting in the energy of the laser photons being shifted up or down. The shift in energy gives information about the vibrational modes in the system. Time-resolved spectroscopy and infrared spectroscopy typically yields similar y ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
 |
MALBAC
Multiple Annealing and Looping Based Amplification Cycles (MALBAC) is a quasilinear whole genome amplification method. Unlike conventional DNA amplification methods that are non-linear or exponential (in each cycle, DNA copied can serve as template for subsequent cycles), MALBAC utilizes special primers that allow amplicons to have complementary ends and therefore to loop, preventing DNA from being copied exponentially. This results in amplification of only the original genomic DNA and therefore reduces amplification bias. MALBAC is “used to create overlapped shotgun amplicons covering most of the genome”.Zong, C.; Lu, S.; Chapman, A.R.; Xie, S. (2012). "Genome-wide detection of single-nucleotide and copy-number variations of a single human cell." ''Science'' 338, 1622. DOI: 10.1126/science.1229164. For next generation sequencing, MALBAC is followed by regular PCR which is used to further amplify amplicons. Technological platform Prior to MALBAC, a single cell is isolate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
Φ29 DNA Polymerase
Φ29 DNA polymerase is an enzyme from the bacteriophage Φ29. It is being increasingly used in molecular biology for multiple displacement DNA amplification procedures, and has a number of features that make it particularly suitable for this application. It was discovered and characterized by Spanish scientists Luis Blanco and Margarita Salas. Φ29 DNA replication Φ29 is a bacteriophage of ''Bacillus subtilis'' with a sequenced, linear, 19,285 base pair DNA genome. Each 5' end is covalently linked to a terminal protein, which is essential in the replication process by acting as a primer for the viral DNA polymerase. A symmetrical mode of replication has been suggested, whereby protein-primed initiation occurs non-simultaneously from either end of the chromosome; this involves two replication origins and two distinct polymerase monomers. Synthesis is continual and involves a strand displacement mechanism. This was demonstrated by the ability of the enzyme to continue to copy t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
 |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
Multiple Displacement Amplification
Multiple displacement amplification (MDA) is a DNA amplification technique. This method can rapidly amplify minute amounts of DNA samples to a reasonable quantity for genomic analysis. The reaction starts by annealing random hexamer Primer (molecular biology), primers to the template: DNA synthesis is carried out by a high fidelity enzyme, preferentially Φ29 DNA polymerase. Compared with conventional Polymerase chain reaction, PCR amplification techniques, MDA does not employ sequence-specific primers but amplifies all DNA, generates larger-sized products with a lower error frequency, and works at a constant temperature. MDA has been actively used in whole genome amplification (WGA) and is a promising method for application to single cell genome sequencing and sequencing-based genetic studies. Background Many biological and forensic cases involving Genetics, genetic analysis require sequencing of DNA from minute amounts of sample, such as DNA from uncultured single cells or trace ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
|
Primer (molecular Biology)
A primer is a short, single-stranded nucleic acid used by all living organisms in the initiation of DNA synthesis. A synthetic primer is a type of oligo, short for oligonucleotide. DNA polymerases (responsible for DNA replication) are only capable of adding nucleotides to the 3’-end of an existing nucleic acid, requiring a primer be bound to the template before DNA polymerase can begin a complementary strand. DNA polymerase adds nucleotides after binding to the RNA primer and synthesizes the whole strand. Later, the RNA strands must be removed accurately and replaced with DNA nucleotides. This forms a gap region known as a nick that is filled in using a ligase. The removal process of the RNA primer requires several enzymes, such as Fen1, Lig1, and others that work in coordination with DNA polymerase, to ensure the removal of the RNA nucleotides and the addition of DNA nucleotides. Living organisms use solely RNA primers, while laboratory techniques in biochemistry and mole ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
|
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research and forensic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
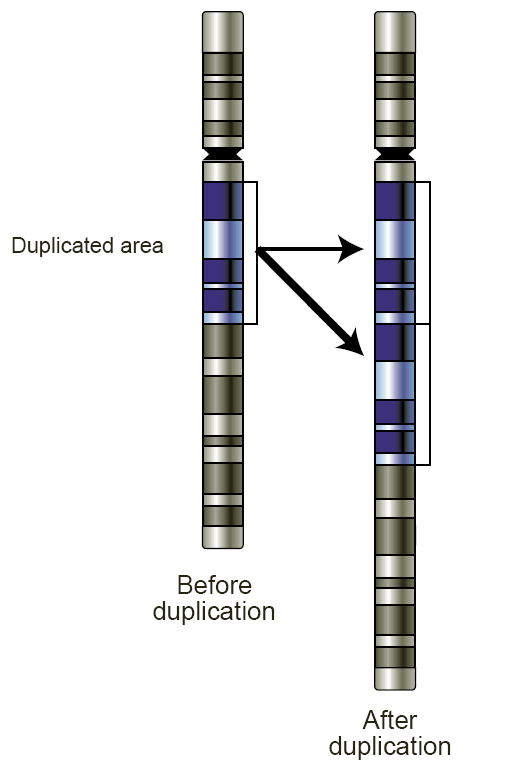 |
Copy Number Variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype. Copy number variations can be generally categorized into two main groups: short repeats and long repeats. However, there are no clear boundaries between the two groups and the classification depends on the nature of the loci of interest. Short repeats include mainly dinucleotide repeats (two repeating nucleotides e.g. A-C-A-C-A-C...) and trinucleotide repeats. Lon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
 |
Single-cell Sequencing
Single-cell sequencing examines the nucleic acid sequence information from individual cells with optimized next-generation sequencing technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment. For example, in cancer, sequencing the DNA of individual cells can give information about mutations carried by small populations of cells. In development, sequencing the RNAs expressed by individual cells can give insight into the existence and behavior of different cell types. In microbial systems, a population of the same species can appear genetically clonal. Still, single-cell sequencing of RNA or epigenetic modifications can reveal cell-to-cell variability that may help populations rapidly adapt to survive in changing environments. Background A typical human cell consists of about 2 x 3.3 billion base pairs of DNA and 600 million mRNA bases. Usually, a mix of millions of cel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
CMOS
Complementary metal–oxide–semiconductor (CMOS, pronounced "sea-moss ", , ) is a type of MOSFET, metal–oxide–semiconductor field-effect transistor (MOSFET) semiconductor device fabrication, fabrication process that uses complementary and symmetrical pairs of p-type semiconductor, p-type and n-type semiconductor, n-type MOSFETs for logic functions. CMOS technology is used for constructing integrated circuit (IC) chips, including microprocessors, microcontrollers, memory chips (including Nonvolatile BIOS memory, CMOS BIOS), and other digital logic circuits. CMOS technology is also used for analog circuits such as image sensors (CMOS sensors), data conversion, data converters, RF circuits (RF CMOS), and highly integrated transceivers for many types of communication. In 1948, Bardeen and Brattain patented an insulated-gate transistor (IGFET) with an inversion layer. Bardeen's concept forms the basis of CMOS technology today. The CMOS process was presented by Fairchild Semico ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
 |
MEMS
MEMS (micro-electromechanical systems) is the technology of microscopic devices incorporating both electronic and moving parts. MEMS are made up of components between 1 and 100 micrometres in size (i.e., 0.001 to 0.1 mm), and MEMS devices generally range in size from 20 micrometres to a millimetre (i.e., 0.02 to 1.0 mm), although components arranged in arrays (e.g., digital micromirror devices) can be more than 1000 mm2. They usually consist of a central unit that processes data (an integrated circuit chip such as microprocessor) and several components that interact with the surroundings (such as microsensors). Because of the large surface area to volume ratio of MEMS, forces produced by ambient electromagnetism (e.g., electrostatic charges and magnetic moments), and fluid dynamics (e.g., surface tension and viscosity) are more important design considerations than with larger scale mechanical devices. MEMS technology is distinguished from molecular nanotechnolo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |