|
Mesquite (software)
Mesquite is a software package primarily designed for phylogenetic analyses. It was developed as a successor to MacClade, when the authors recognized that implementing a modular architecture in MacClade would be infeasible. Mesquite is largely written in Java and uses NEXUS-formatted files as input. Mesquite is available as a compiled executable for Macintosh, Windows, and Unix-like platforms, and the source code is available on GitHub. Rationale The architecture of Mesquite was explicitly designed to exploit the modularity and extensibility of Object-oriented programming. In practice, Mesquite ''modules'' are Java classes, usually concrete sub-classes of abstract class definitions. This modular architecture has afforded the development of additional packages that can be used as plug-ins to Mesquite. Scope Mesquite provides a number of analyses including ancestral state estimation and diversification analysis. Mesquite also contains tools for simulating data such as species ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Java (programming Language)
Java is a High-level programming language, high-level, General-purpose programming language, general-purpose, Memory safety, memory-safe, object-oriented programming, object-oriented programming language. It is intended to let programmers ''write once, run anywhere'' (Write once, run anywhere, WORA), meaning that compiler, compiled Java code can run on all platforms that support Java without the need to recompile. Java applications are typically compiled to Java bytecode, bytecode that can run on any Java virtual machine (JVM) regardless of the underlying computer architecture. The syntax (programming languages), syntax of Java is similar to C (programming language), C and C++, but has fewer low-level programming language, low-level facilities than either of them. The Java runtime provides dynamic capabilities (such as Reflective programming, reflection and runtime code modification) that are typically not available in traditional compiled languages. Java gained popularity sh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Unix-like
A Unix-like (sometimes referred to as UN*X, *nix or *NIX) operating system is one that behaves in a manner similar to a Unix system, although not necessarily conforming to or being certified to any version of the Single UNIX Specification. A Unix-like Application software, application is one that behaves like the corresponding List of POSIX commands, Unix command or Unix shell, shell. Although there are general Unix philosophy, philosophies for Unix design, there is no technical standard defining the term, and opinions can differ about the degree to which a particular operating system or application is Unix-like. Some well-known examples of Unix-like operating systems include Linux, FreeBSD and OpenBSD. These systems are often used on servers as well as on personal computers and other devices. Many popular applications, such as the Apache HTTP Server, Apache web server and the Bash (Unix shell), Bash shell, are also designed to be used on Unix-like systems. Definition The Open ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cross-platform
Within computing, cross-platform software (also called multi-platform software, platform-agnostic software, or platform-independent software) is computer software that is designed to work in several Computing platform, computing platforms. Some cross-platform software requires a separate build for each platform, but some can be directly run on any platform without special preparation, being written in an interpreted language or compiled to portable bytecode for which the Interpreter (computing), interpreters or run-time packages are common or standard components of all supported platforms. For example, a cross-platform application software, application may run on Linux, macOS and Microsoft Windows. Cross-platform software may run on many platforms, or as few as two. Some frameworks for cross-platform development are Codename One, ArkUI-X, Kivy (framework), Kivy, Qt (software), Qt, GTK, Flutter (software), Flutter, NativeScript, Xamarin, Apache Cordova, Ionic (mobile app framework ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nexus File
The extensible NEXUS file format is widely used in phylogenetics, evolutionary biology, and bioinformatics. It stores information about taxa In biology, a taxon (back-formation from ''taxonomy''; : taxa) is a group of one or more populations of an organism or organisms seen by taxonomists to form a unit. Although neither is required, a taxon is usually known by a particular name and ..., morphological character states, DNA and protein sequence alignments, distances, and phylogenetic trees. The NEXUS format also allows the storage of data that can facilitate analyses, such as sets of characters or taxa. Many popular phylogenetic programs, including PAUP*,PAUP* — Phylogenetic Analysis Using Parsimony *and other methods [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Systematic Biology
Systematics is the study of the diversification of living forms, both past and present, and the relationships among living things through time. Relationships are visualized as evolutionary trees (synonyms: phylogenetic trees, phylogenies). Phylogenies have two components: branching order (showing group relationships, graphically represented in cladograms) and branch length (showing amount of evolution). Phylogenetic trees of species and higher taxa are used to study the evolution of traits (e.g., anatomical or molecular characteristics) and the distribution of organisms (biogeography). Systematics, in other words, is used to understand the evolutionary history of life on Earth. The word systematics is derived from the Latin word of Ancient Greek origin '' systema,'' which means systematic arrangement of organisms. Carl Linnaeus used 'Systema Naturae' as the title of his book. Branches and applications In the study of biological systematics, researchers use the different branch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GitHub
GitHub () is a Proprietary software, proprietary developer platform that allows developers to create, store, manage, and share their code. It uses Git to provide distributed version control and GitHub itself provides access control, bug tracking system, bug tracking, software feature requests, task management, continuous integration, and wikis for every project. Headquartered in California, GitHub, Inc. has been a subsidiary of Microsoft since 2018. It is commonly used to host open source software development projects. GitHub reported having over 100 million developers and more than 420 million Repository (version control), repositories, including at least 28 million public repositories. It is the world's largest source code host Over five billion developer contributions were made to more than 500 million open source projects in 2024. About Founding The development of the GitHub platform began on October 19, 2005. The site was launched in April 2008 by Tom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Object-oriented Programming
Object-oriented programming (OOP) is a programming paradigm based on the concept of '' objects''. Objects can contain data (called fields, attributes or properties) and have actions they can perform (called procedures or methods and implemented in code). In OOP, computer programs are designed by making them out of objects that interact with one another. Many of the most widely used programming languages (such as C++, Java, and Python) support object-oriented programming to a greater or lesser degree, typically as part of multiple paradigms in combination with others such as imperative programming and declarative programming. Significant object-oriented languages include Ada, ActionScript, C++, Common Lisp, C#, Dart, Eiffel, Fortran 2003, Haxe, Java, JavaScript, Kotlin, Logo, MATLAB, Objective-C, Object Pascal, Perl, PHP, Python, R, Raku, Ruby, Scala, SIMSCRIPT, Simula, Smalltalk, Swift, Vala and Visual Basic.NET. History The idea of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Class (computer Programming)
In object-oriented programming, a class defines the shared aspects of objects created from the class. The capabilities of a class differ between programming languages, but generally the shared aspects consist of state ( variables) and behavior ( methods) that are each either associated with a particular object or with all objects of that class. Object state can differ between each instance of the class whereas the class state is shared by all of them. The object methods include access to the object state (via an implicit or explicit parameter that references the object) whereas class methods do not. If the language supports inheritance, a class can be defined based on another class with all of its state and behavior plus additional state and behavior that further specializes the class. The specialized class is a ''sub-class'', and the class it is based on is its ''superclass''. Attributes Object lifecycle As an instance of a class, an object is constructed from a class via '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ancestral Reconstruction
Ancestral reconstruction (also known as ''Character Mapping'' or ''Character Optimization'') is the extrapolation back in time from measured characteristics of individuals, populations, or species to their Common descent, common ancestors. It is an important application of phylogenetics, the reconstruction and study of the evolutionary relationships among individuals, populations or species to their ancestors. In the context of evolutionary biology, ancestral reconstruction can be used to recover different kinds of ancestral character states of organisms that lived millions of years ago. These states include the Nucleic acid sequence, genetic sequence (ancestral sequence reconstruction), the Peptide sequence, amino acid sequence of a protein, the composition of a genome (e.g., gene order), a measurable characteristic of an organism (phenotype), and the species distribution, geographic range of an ancestral population or species (ancestral range reconstruction). This is desirable beca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
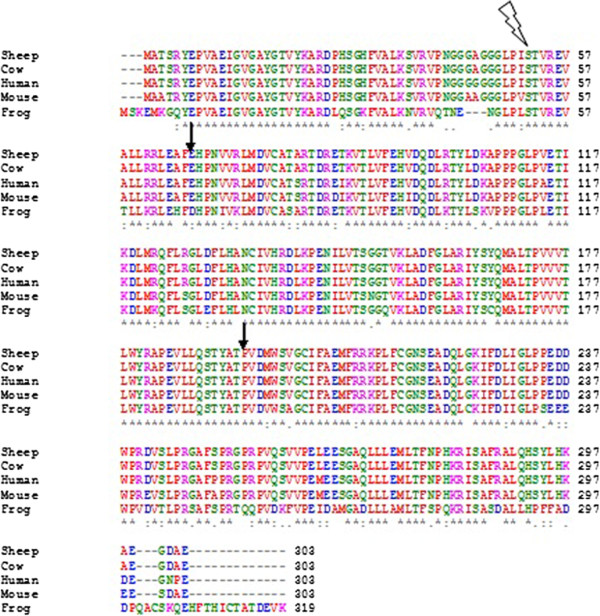
Multiple Sequence Alignment
Multiple sequence alignment (MSA) is the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. These alignments are used to infer evolutionary relationships via phylogenetic analysis and can highlight homologous features between sequences. Alignments highlight mutation events such as point mutations (single amino acid or nucleotide changes), insertion mutations and deletion mutations, and alignments are used to assess sequence conservation and infer the presence and activity of protein domains, tertiary structures, secondary structures, and individual amino acids or nucleotides. Multiple sequence alignments require more sophisticated methodologies than pairwise alignments, as they are more computationally complex. Most multiple sequence alignment programs use heuristic methods rather than global optimization because identifying the optimal alignment between more than a few sequences of moderate length is prohibiti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clustal
Clustal is a computer program used for multiple sequence alignment in bioinformatics. The software and its algorithms have gone through several iterations, with ClustalΩ (Omega) being the latest version . It is available as standalone software, via a Clustal#External links, web interface, and through a server hosted by the European Bioinformatics Institute. Clustal has been an important bioinformatic software, with two of its academic publications amongst the top 100 papers cited of all time, according to Nature (journal), Nature in 2014. History Version history * Clustal: The original software for multiple sequence alignments, created by Desmond G. Higgins, Des Higgins in 1988, was based on deriving a guide tree from pairwise sequences of Amino acid, amino acids or Nucleotide, nucleotides. * ClustalV: The second generation of Clustal, released in 1992. It introduced the ability to create new alignments from existing alignments in a process known as phylogenetic tree re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |