|
MBNL1
Muscleblind Like Splicing Regulator 1 (MBNL1) is an RNA splicing protein that in humans is encoded by the ''MBNL1'' gene. It has a well characterized role in Myotonic dystrophy where impaired splicing disrupts muscle development and function. In addition to regulating mRNA maturation of hundreds of genes MBNL1 (along with its paralogs MBNL2 & MBNL3) autoregulate alternative splicing of the MBNL1 pre-mRNA transcript. The founding member of the human MBNL family of proteins was the Drosophila Muscleblind protein (PMID 9334280). Human MBNL1 is an alternative splicing regulator that harbors dual function as both a repressor and activator for terminal muscle differentiation. The repressive function of Human MBNL1 by sequestering at normal splice sites has been shown to lead to RNA-splicing defects that lead to muscular diseases. The gene can be alternatively spliced into multiple functionally distinct isoforms, some of which linked to be involved in cancer biology. Human MBNL1 is a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Myotonic Dystrophy
Myotonic dystrophy (DM) is a type of muscular dystrophy, a group of genetic disorders that cause progressive muscle loss and weakness. In DM, muscles are often unable to relax after contraction. Other manifestations may include cataracts, intellectual disability and heart conduction problems. In men, there may be early balding and an inability to have children. While myotonic dystrophy can occur at any age, onset is typically in the 20s and 30s. Myotonic dystrophy is caused by a genetic mutation in one of two genes. Mutation of the '' DMPK'' gene causes myotonic dystrophy type 1 (DM1). Mutation of '' CNBP'' gene causes type 2 (DM2). DM is typically inherited from a person's parents, following an autosomal dominant inheritance pattern, and it generally worsens with each generation. A type of DM1 may be apparent at birth. DM2 is generally milder. Diagnosis is confirmed by genetic testing. There is no cure. Treatments may include braces or wheelchairs, pacemakers and non-i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
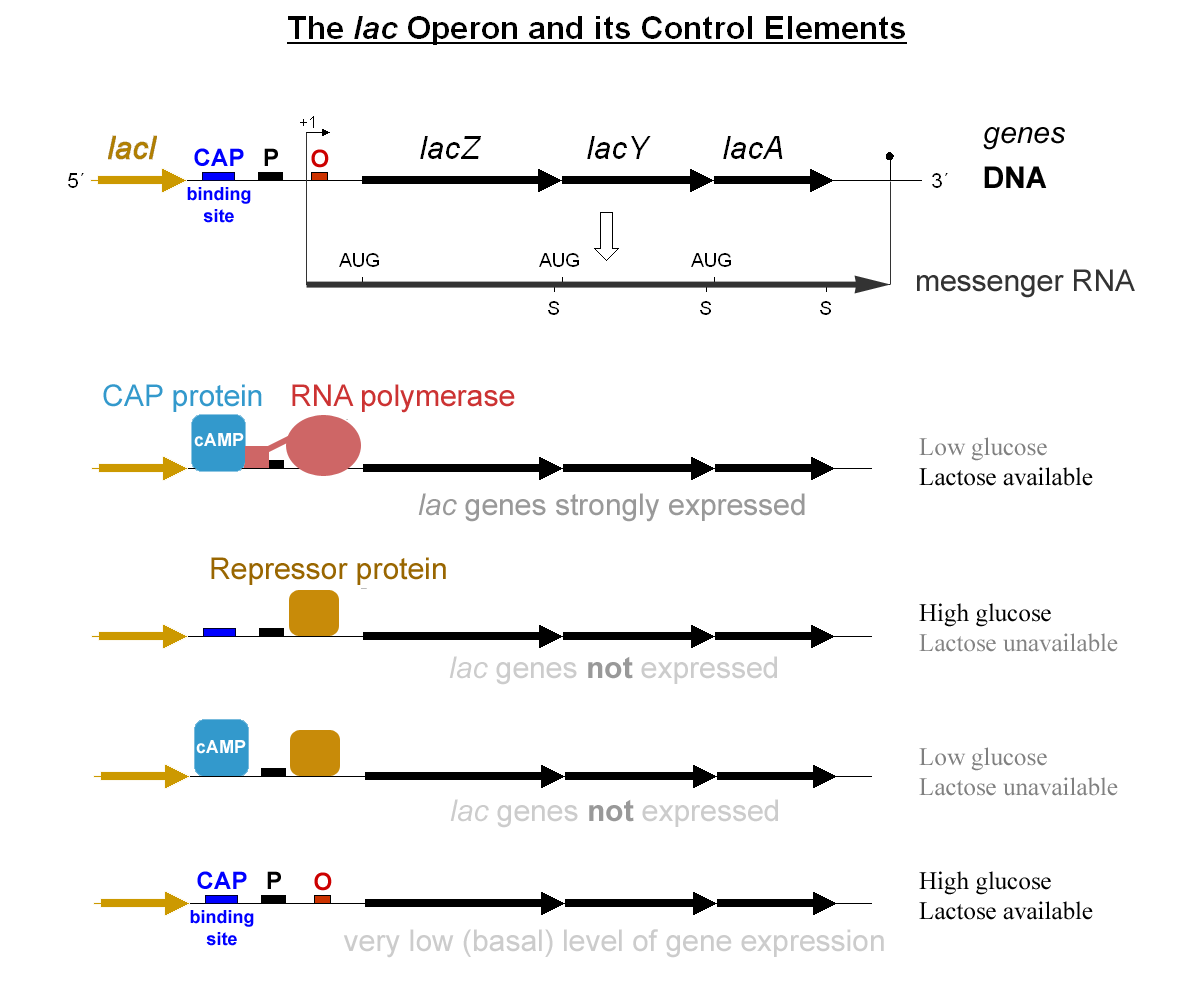
RNA Splicing
RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing all the introns (non-coding regions of RNA) and ''splicing'' back together exons (coding regions). For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, splicing and translation is called gene expression, the central dogma of molecular biology. Splicing pathways Several methods of RNA spl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid resid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Splicing Regulatory Element
Splicing regulatory element (SRE) are ''cis''-acting sequences in pre-mRNA, which either enhance or silence (suppress) the splicing of introns, or in general regulates the constitutive or alternative splicing of this pre-mRNA. SREs recruit ''trans''-acting splicing factors to activate or suppress the splice site recognition or spliceosome assembly. The "context dependence" of SREs is categorized into at least two studied groups: (a) the location-dependent activity of SREs: the activity varies with the relative positions of SREs in pre-mRNA; (b) the gene-dependent activity of SREs: the SRE activity observed in one gene is lost when the SRE is moved to another gene. SREs are: :present in exons: exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs) :present in introns: intronic splicing enhancers (ISEs), intronic splicing silencer An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is deriv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repressor
In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA. An RNA-binding repressor binds to the mRNA and prevents translation of the mRNA into protein. This blocking or reducing of expression is called repression. Function If an inducer, a molecule that initiates the gene expression, is present, then it can interact with the repressor protein and detach it from the operator. RNA polymerase then can transcribe the message (expressing the gene). A co-repressor is a molecule that can bind to the repressor and make it bind to the operator tightly, which decreases transcription. A repressor that binds with a co-repressor is termed an ''aporepressor'' or ''inactive repressor''. One type of aporepressor is the trp repre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activator (genetics)
A transcriptional activator is a protein ( transcription factor) that increases transcription of a gene or set of genes. Activators are considered to have ''positive'' control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain". Most activators function by binding sequence-specifically to a regulatory DNA site located near a promoter and making protein–protein interactions with the general transcription machinery (RNA polymerase and general transcription factors), thereby facilitating the binding of the general transcription machinery to the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Splice Site Mutation
A splice site mutation is a genetic mutation that inserts, deletes or changes a number of nucleotides in the specific site at which splicing takes place during the processing of precursor messenger RNA into mature messenger RNA. Splice site consensus sequences that drive exon recognition are located at the very termini of introns. The deletion of the splicing site results in one or more introns remaining in mature mRNA and may lead to the production of abnormal proteins. When a splice site mutation occurs, the mRNA transcript possesses information from these introns that normally should not be included. Introns are supposed to be removed, while the exons are expressed. The mutation must occur at the specific site at which intron splicing occurs: within non-coding sites in a gene, directly next to the location of the exon. The mutation can be an insertion, deletion, frameshift, etc. The splicing process itself is controlled by the given sequences, known as splice-donor and splic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |