|
Linker Histone H1 Variants
In molecular biology, the linker histone H1 is a protein family forming a critical component of eukaryotic chromatin. H1 histones bind to the linker DNA exiting from the nucleosome core particle, while the core histones ( H2A, H2B, H3 and H4) form the octamer core of the nucleosome around which the DNA is wrapped. H1 forms a complex family of related proteins with distinct specificity for tissues, developmental stages, and organisms in which they are expressed. Individual H1 proteins are often referred to as '' isoforms'' or ''variants''. The discovery of H1 variants in calf thymus preceded the discovery of core histone variants. Human linker histone variants In human and mouse cells 11 H1 variants have been described and are encoded by single genes. Six of the variants are mainly expressed during the S phase and hence replication-dependent. They are encoded by genes within histone cluster 1 located in human cells on chromosome 6. The five further variants are expr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H1 Binding To The Nucleosome
H1, H-1, H01, H I may refer to: Places *Interstate H-1, a highway in Hawaii * Area H1, the area of Hebron controlled by the Palestinian Authority under the Hebron Protocol Science * H1 (particle detector) * Histamine H1 receptor * Histone H1, a protein * British NVC community H1, a heath zone * Hydrogen atom (H1) * Protium (isotope), an isotope of hydrogen * H I region, a cloud in the interstellar medium * ATC code H01 ''Pituitary and hypothalamic hormones and analogues'', a subgroup of the Anatomical Therapeutic Chemical Classification System * Haplogroup H1 (other), a grouping in genetics based on certain similarities * Alternative hypothesis (H1) Technology * , level 1 heading markup for HTML Web pages; see HTML element * DSC-H1, a Sony Cybershot digital camera * H1, John Harrison's first marine chronometer * Tianwen-1 (formerly ''Huoxing-1''), first Chinese space probe to reach Mars * H-1NF, the Australian Plasma Fusion Research Facility, formerly H-1 Heliac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H2B
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes. Structure Histone H2B is a lightweight structural protein made of 126 amino acids. Many of these amino acids have a positive charge at cellular pH, which allows them to interact with the negatively charged phosphate groups in DNA. Along with a central globular domain, histone H2B has two flexible histone tails that extend outwards – one at the N-terminal end and one at C-terminal end. These are highly involved in condensing chromatin from the beads-on-a-string conformation to a 30-nm fiber. Similar to other histone proteins, histone H2B has a distinct histone fold that is optimized for histone-histone as well as histone-DNA interactions. Two copies of histone H2B come together with two copies each of histone H2A, histone H3, and hi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetics & Chromatin
''Epigenetics & Chromatin'' is a peer-reviewed open access scientific journal published by BioMed Central that covers the biology of epigenetics and chromatin. Scope ''Epigenetics & Chromatin'' is a peer-reviewed open access scientific journal that publishes research related to epigenetic inheritance and chromatin-based interactions. First published in 2008 by BioMed Central, its overall aim is to understand the regulation of gene and chromosomal elements during the processes of cell division, cell differentiation, and any alterations in the environment. To date, 13 volumes have been published. Usage As of October 2020, there has been over 340,000 downloads and over 750 Altmetric mentions. Metrics Impact factor According to Journal Citation Reports, it received an impact factor of 4.237 in 2019. Its current SCImago Journal Rank is 2.449. Citation impact Its 2-year and 5-year citation impact factor is 4.237 and 4.763, respectively. Its Source Normalized Impact per Pap ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Linker Histone Variant Evolutionary Tree
Linker or linkers may refer to: Computing * Linker (computing), a computer program that takes one or more object files generated by a compiler or generated by an assembler and links them with libraries, generating an executable program or shared library ** GNU linker, the classic GNU Project's implementation of the Unix linker command ld * Dynamic linker, the part of an operating system that loads and links the shared libraries for an executable program at run time People * Amy Linker (born 1966), American actress * Zita Linker (1917–2009), Israeli politician * Eduard Linkers (1912–2004), an Austrian actor Biology * Linker DNA, the part of a genomic DNA strand that connects two nucleosomes * Polylinker or multiple cloning site, a short segment of DNA with many restriction sites * Signal transducing adaptor protein, proteins that provide mechanisms by which receptors can amplify and regulate downstream effector proteins ** Linker of activated T cells, a protein in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isoforms
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions genes revealed by the human genome project and the large diversity of proteins seen in an organism: differen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Octamer
A histone octamer is the eight-protein complex found at the center of a nucleosome core particle. It consists of two copies of each of the four core histone proteins ( H2A, H2B, H3, and H4). The octamer assembles when a tetramer, containing two copies of H3 and two of H4, complexes with two H2A/H2B dimers. Each histone has both an N-terminal tail and a C-terminal histone-fold. Each of these key components interacts with DNA in its own way through a series of weak interactions, including hydrogen bonds and salt bridges. These interactions keep the DNA and the histone octamer loosely associated, and ultimately allow the two to re-position or to separate entirely. History of research Histone post-translational modifications were first identified and listed as having a potential regulatory role on the synthesis of RNA in 1964. Since then, over several decades, chromatin theory has evolved. Chromatin subunit models as well as the notion of the nucleosome were established in 197 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H4
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H4 is involved with the structure of the nucleosome of the 'beads on a string' organization. Histone proteins are highly post-translationally modified. Covalently bonded modifications include acetylation and methylation of the N-terminal tails. These modifications may alter expression of genes located on DNA associated with its parent histone octamer. Histone H4 is an important protein in the structure and function of chromatin, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Genetics Histone H4 is encoded in multiple genes at different loci including: HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L, HIST2H4A, HIST2H4B, HIST4H4. Evolution Histone ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure. Histone proteins are highly post-translationally modified however Histone H3 is the most extensively modified of the five histones. The term "Histone H3" alone is purposely ambiguous in that it does not distinguish between sequence variants or modification state. Histone H3 is an important protein in the emerging field of epigenetics, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Epigenetics and post-translational modifications The N-terminus of H3 protrudes from the globular nucleosome core and is susceptible to post-translational modification that influence cellular processes. These modifications include the covalent attachment ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H2A
Histone H2A is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. The other histone proteins are: H1, H2B, H3 and H4. Background Histones are proteins that package DNA into nucleosomes. Histones are responsible for maintaining the shape and structure of a nucleosome. One chromatin molecule is composed of at least one of each core histones per 100 base pairs of DNA. There are five families of histones known to date; these histones are termed H1/H5, H2A, H2B, H3, and H4. H2A is considered a core histone, along with H2B, H3 and H4. Core formation first occurs through the interaction of two H2A molecules. Then, H2A forms a dimer with H2B; the core molecule is complete when H3-H4 also attaches to form a tetramer. Sequence variants Histone H2A is composed of non-allelic variants. The term "Histone H2A" is intentionally non-specific and refers to a variety of closely related proteins that vary often by only a few amino acids. Ap ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wrapping of DNA around his ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
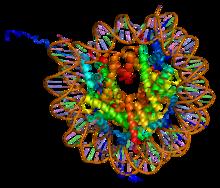
Nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4. DNA must be compacted into nucleosomes to fit within the cell nucleus. In addition to nucleosome wrapping, eukaryotic chromatin is further compacted by being folded into a series of more complex structures, eventually forming a chromosome. Each human cell contains about 30 million nucleosomes. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones. Nucleosome positions in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |