|
Ligase
In biochemistry, a ligase is an enzyme that can catalyze the joining (ligation) of two molecules by forming a new chemical bond. This is typically via hydrolysis of a small pendant chemical group on one of the molecules, typically resulting in the formation of new C-O, C-S, or C-N bonds. For example, DNA ligase can join two complementary fragments of nucleic acid by forming phosphodiester bonds, and repair single stranded breaks that arise in double stranded DNA during replication. In general, a ligase catalyzes the following dehydration reaction, thus joining molecules A and B: A-OH + B-H → A–B + H2O Nomenclature The naming of ligases is inconsistent and so these enzymes are commonly known by several different names. Generally, the common names of ligases include the word "ligase", such as in DNA ligase, an enzyme commonly used in molecular biology laboratories to join together DNA fragments. However, many common names use the term "synthetase" or "synthase" instead, beca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biochemistry
Biochemistry, or biological chemistry, is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology, and metabolism. Over the last decades of the 20th century, biochemistry has become successful at explaining living processes through these three disciplines. Almost all List of life sciences, areas of the life sciences are being uncovered and developed through biochemical methodology and research.#Voet, Voet (2005), p. 3. Biochemistry focuses on understanding the chemical basis that allows biomolecule, biological molecules to give rise to the processes that occur within living Cell (biology), cells and between cells,#Karp, Karp (2009), p. 2. in turn relating greatly to the understanding of tissue (biology), tissues and organ (anatomy), organs as well as organism structure and function.#Miller, Miller (2012). p. 62. Biochemistry is closely ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chelatase
In biochemistry, chelatases are enzymes that catalyze the insertion ("metalation") of naturally occurring tetrapyrroles. Many tetrapyrrole-based cofactors exist in nature including hemes, chlorophylls, and vitamin B12. These metallo cofactors are derived by the reaction of metal cations with tetrapyrroles, which are not ligands ''per se'', but the conjugate acids thereof. In the case of ferrochelatases, the reaction that chelatases catalyze is: :Fe2+ + H2P → FeP + 2 H+ In the above equation H2P represents a sirohydrochlorin or a porphyrin, such as protoporphyrin IX. Chelatases are required because porphyrins and related macrocyclic ligands are extremely slow to metalate, despite favorable thermodynamics. These low rates are attributed to the tight fit of the metal into the rigid 18- or 17-membered tetrapyrrole macrocycle. Several families of chelatase are known including cobalt chelatase, magnesium chelatase, and ferrochelatase. Nickel insertion into a sirohydrochlori ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin Ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another protein (the substrate) by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalysis, catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks Covalent bond, bonds. Proteases are involved in numerous biological pathways, including Digestion#Protein digestion, digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently convergent evolution, evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Classification Based on catalytic residue Proteases can be classified into seven broad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclease
In biochemistry, a nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids. Nucleases variously affect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning. There are two primary classifications based on the locus of activity. Exonucleases digest nucleic acids from the ends. Endonucleases act on regions in the ''middle'' of target molecules. They are further subcategorized as deoxyribonucleases and ribonucleases. The former acts on DNA, the latter on RNA. History In the late 1960s, scientists Stuart Linn and Werner Arber isolated examples of the two types of enzymes responsible for phage growth restriction in Escherichi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
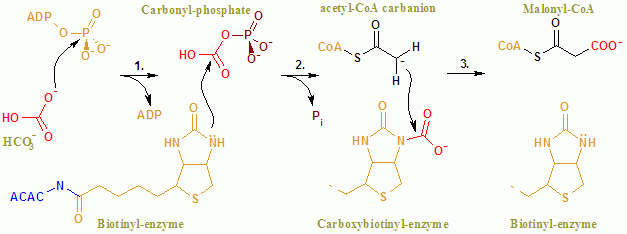
Acetyl-CoA Carboxylase
Acetyl-CoA carboxylase (ACC) is a biotin-dependent enzyme () that catalyzes the irreversible carboxylation of acetyl-CoA to produce malonyl-CoA through its two catalytic activities, biotin carboxylase (BC) and carboxyltransferase (CT). ACC is a multi-subunit enzyme in most prokaryotes and in the chloroplasts of most plants and algae, whereas it is a large, multi-domain enzyme in the cytoplasm of most eukaryotes. The most important function of ACC is to provide the malonyl-CoA substrate for the biosynthesis of fatty acids. The activity of ACC can be controlled at the transcriptional level as well as by small molecule modulators and covalent modification. The human genome contains the genes for two different ACCs—'' ACACA'' and '' ACACB''. Structure Prokaryotes and plants have multi-subunit ACCs composed of several polypeptides. Biotin carboxylase (BC) activity, biotin carboxyl carrier protein (BCCP), and carboxyl transferase (CT) activity are each contained on a differ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
-ase
The suffix -ase is used in biochemistry to form names of enzymes. The most common way to name enzymes is to add this suffix onto the end of the substrate, ''e.g.'' an enzyme that breaks down peroxides may be called peroxidase; the enzyme that produces telomeres is called telomerase. Sometimes enzymes are named for the function they perform, rather than substrate, e.g. the enzyme that polymerizes (assembles) DNA into strands is called polymerase; see also reverse transcriptase. Etymology The ''-ase'' suffix is a libfix derived from " diastase", the first recognized enzyme. Its usage in subsequently discovered enzymes was proposed by Émile Duclaux, with the intention of honoring the first scientists to isolate diastase. See also *Amylase *DNA polymerase A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually wo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Latin Language
Latin ( or ) is a classical language belonging to the Italic languages, Italic branch of the Indo-European languages. Latin was originally spoken by the Latins (Italic tribe), Latins in Latium (now known as Lazio), the lower Tiber area around Rome, Italy. Through the expansion of the Roman Republic, it became the dominant language in the Italian Peninsula and subsequently throughout the Roman Empire. It has greatly influenced many languages, Latin influence in English, including English, having contributed List of Latin words with English derivatives, many words to the English lexicon, particularly after the Christianity in Anglo-Saxon England, Christianization of the Anglo-Saxons and the Norman Conquest. Latin Root (linguistics), roots appear frequently in the technical vocabulary used by fields such as theology, List of Latin and Greek words commonly used in systematic names, the sciences, List of medical roots, suffixes and prefixes, medicine, and List of Latin legal terms ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Classical Compound
Neoclassical compounds are compound words composed from combining forms (which act as affixes or stems) derived from Classical_language#Classical_studies, classical languages (classical Latin or ancient Greek) root (linguistics), roots. Neo-Latin comprises many such words and is a substantial component of the technology, technical and science, scientific lexicon of English language, English and other languages, via international scientific vocabulary (ISV). For example, Greek ''wikt:bio-#Prefix, bio-'' combines with ''wikt:-graphy#Suffix, -graphy'' to form ''biography'' ("life" + "writing/recording"). Source of international technical vocabulary Neoclassical compounds represent a significant source of Neo-Latin vocabulary. Moreover, since these words are composed from classical languages whose prestige is or was respected throughout the Western European culture, these words typically appear in many different languages. Their widespread use makes technical writing generally acces ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin Ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another protein (the substrate) by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Membranome Database
Membranome database provides structural and functional information about more than 6000 single-pass (bitopic) transmembrane proteins from ''Homo sapiens'', ''Arabidopsis thaliana'', ''Dictyostelium discoideum'', ''Saccharomyces cerevisiae'', ''Escherichia coli'' and ''Methanocaldococcus jannaschii''. Bitopic membrane proteins consist of a single transmembrane alpha-helix connecting water-soluble domains of the protein situated at the opposite sides of a biological membrane. These proteins are frequently involved in the signal transduction and communication between cells in multicellular organisms. The database provides information about the individual proteins including computationally generated three-dimensional models of their transmembrane alpha-helices spatially arranged in the membrane, topology, intracellular localizations, amino acid sequences, domain architecture, functional annotation and available experimental structures from the Protein Data Bank. It also provides ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |