|
Isu1
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial is a protein that in humans is encoded by the ''ISCU'' gene. It encodes an iron-sulfur (Fe-S) cluster scaffold protein involved in 2Fe-2S">2Fe-2S_cluster.html" ;"title="nowiki/>2Fe-2S cluster">2Fe-2Sand [4Fe-4S] cluster synthesis and maturation. A deficiency of ISCU is associated with a mitochondrial myopathy with lifelong exercise intolerance where only minor exertion causes tachycardia, shortness of breath, muscle weakness and myalgia. Updated 2011 Sep 1 Structure ''ISCU'' is located on the q arm of chromosome 12 in position 23.3 and has 8 exons. ISCU, the protein encoded by this gene, is a member of the NifU family. It is an iron-sulfur transferase that contains binding sites for 2Fe-2S">2Fe-2S_cluster.html" ;"title="nowiki/>2Fe-2S cluster">2Fe-2Sand [4Fe-4S] clusters. ISCU contains a transit peptide, 4 Beta sheet, beta strands, 4 alpha helixes, and 4 turns. Alternative splicing results in transcript variants encoding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid resid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transit Peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-terminus (or occasionally nonclassically at the C-terminus or internally) of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles (the endoplasmic reticulum, Golgi or endosomes), secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide. Function (translocation) Signal peptides function to prompt a cell to transloc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GLRX5
Glutaredoxin 5, also known as GLRX5, is a protein which in humans is encoded by the ''GLRX5'' gene located on chromosome 14. This gene encodes a mitochondrial protein, which is evolutionarily conserved. It is involved in the biogenesis of iron- sulfur clusters, which are required for normal iron homeostasis. Mutations in this gene are associated with autosomal recessive pyridoxine-refractory sideroblastic anemia. Structure The GLRX5 gene contains 2 exons and encodes for a protein that is 13 kDa in size. The protein is highly expressed in erythroid cells. Crystal structure of the GLRX5 protein reveals that the protein likely exists as a tetramer with two Fe-S clusters buried in the interior. Function GLRX5 is a mitochondrial protein is conserved evolutionarily and plays a role in the formation of iron-sulfur clusters, which function to maintain iron homeostasis within the mitochondria and in the cell. GLRX5 is required for the steps in haem synthesis that involves mi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
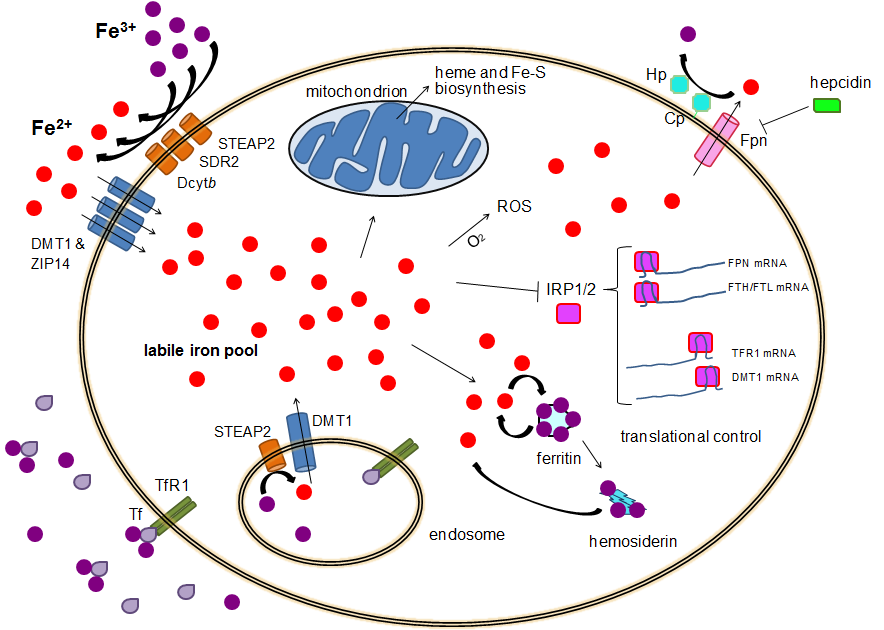
Human Iron Metabolism
Human iron metabolism is the set of chemical reactions that maintain human homeostasis of iron at the systemic and cellular level. Iron is both necessary to the body and potentially toxic. Controlling iron levels in the body is a critically important part of many aspects of human health and disease. Hematologists have been especially interested in systemic iron metabolism because iron is essential for red blood cells, where most of the human body's iron is contained. Understanding iron metabolism is also important for understanding diseases of iron overload, such as hereditary hemochromatosis, and iron deficiency, such as iron-deficiency anemia. Importance of iron regulation Iron is an essential bioelement for most forms of life, from bacteria to mammals. Its importance lies in its ability to mediate electron transfer. In the ferrous state (Fe2+), iron acts as an electron donor, while in the ferric state (Fe3+) it acts as an acceptor. Thus, iron plays a vital role in the catalys ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metabolism
Metabolism (, from el, μεταβολή ''metabolē'', "change") is the set of life-sustaining chemical reactions in organisms. The three main functions of metabolism are: the conversion of the energy in food to energy available to run cellular processes; the conversion of food to building blocks for proteins, lipids, nucleic acids, and some carbohydrates; and the elimination of metabolic wastes. These enzyme-catalyzed reactions allow organisms to grow and reproduce, maintain their structures, and respond to their environments. The word metabolism can also refer to the sum of all chemical reactions that occur in living organisms, including digestion and the transportation of substances into and between different cells, in which case the above described set of reactions within the cells is called intermediary (or intermediate) metabolism. Metabolic reactions may be categorized as '' catabolic'' – the ''breaking down'' of compounds (for example, of glucose to pyruvate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are Ribozyme, catalytic RNA molecules, called ribozymes. Enzymes' Chemical specificity, specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound. Cofactors can be divided into two types: inorganic ions and complex organic molecules called coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Note that some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a "prosthetic group", which consists of a coenzyme that is tightly (or even covalently) and permanently bound to a prote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome 1
Chromosome 1 is the designation for the largest human chromosome. Humans have two copies of chromosome 1, as they do with all of the autosomes, which are the non- sex chromosomes. Chromosome 1 spans about 249 million nucleotide base pairs, which are the basic units of information for DNA.http://vega.sanger.ac.uk/Homo_sapiens/mapview?chr=1 Chromosome size and number of genes derived from this database, retrieved 2012-03-11. It represents about 8% of the total DNA in human cells. It was the last completed chromosome, sequenced two decades after the beginning of the Human Genome Project. Genes Number of genes The following are some of the gene count estimates of human chromosome 1. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project ( CCDS) takes an extremely conservative str ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudogene
Pseudogenes are nonfunctional segments of DNA that resemble functional genes. Most arise as superfluous copies of functional genes, either directly by DNA duplication or indirectly by reverse transcription of an mRNA transcript. Pseudogenes are usually identified when genome sequence analysis finds gene-like sequences that lack regulatory sequences needed for transcription or translation, or whose coding sequences are obviously defective due to frameshifts or premature stop codons. Most non-bacterial genomes contain many pseudogenes, often as many as functional genes. This is not surprising, since various biological processes are expected to accidentally create pseudogenes, and there are no specialized mechanisms to remove them from genomes. Eventually pseudogenes may be deleted from their genomes by chance DNA replication or DNA repair errors, or they may accumulate so many mutational changes that they are no longer recognizable as former genes. Analysis of these degeneration ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondrion
A mitochondrion (; ) is an organelle found in the cells of most Eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'' was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase coined by Philip Siekevitz in a 1957 article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). A large number of unicellular organisms, such as microsporidia, parabasalids and diplomonads, have reduced or transformed their mitochondria into other structures. One eukaryote, '' Monocercomonoides'', is known to have completely lost its mitochondria, and one multicellular organ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
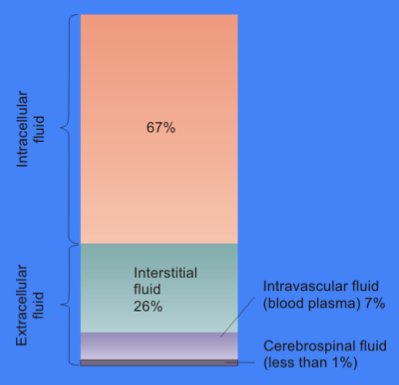
Cytosol
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells ( intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into many compartments. In the eukaryotic cell, the cytosol is surrounded by the cell membrane and is part of the cytoplasm, which also comprises the mitochondria, plastids, and other organelles (but not their internal fluids and structures); the cell nucleus is separate. The cytosol is thus a liquid matrix around the organelles. In prokaryotes, most of the chemical reactions of metabolism take place in the cytosol, while a few take place in membranes or in the periplasmic space. In eukaryotes, while many metabolic pathways still occur in the cytosol, others take place within organelles. The cytosol is a complex mixture of substances dissolved in water. Although water forms the large majority of the cytosol, its structure and prop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Isoform
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions genes revealed by the human genome project and the large diversity of proteins seen in an organism: differen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |