|
Intergenic Region
An intergenic region is a stretch of DNA sequences located between genes. Intergenic regions may contain functional elements and junk DNA. Properties and functions Intergenic regions may contain a number of functional DNA sequences such as promoters and regulatory elements, enhancers, spacers, and (in eukaryotes) centromeres. They may also contain origins of replication, scaffold attachment regions, and transposons and viruses. Non-functional DNA elements such as pseudogenes and repetitive DNA, both of which are types of junk DNA, can also be found in intergenic regions—although they may also be located within genes in introns. It is possible that these regions contain as of yet unidentified functional elements, such as non-coding genes or regulatory sequences. This indeed occurs occasionally, but the amount of functional DNA discovered usually constitute only a tiny fraction of the overall amount of intergenic or intronic DNA. Intergenic regions in differ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes. During gene expression (the synthesis of Gene product, RNA or protein from a gene), DNA is first transcription (biology), copied into RNA. RNA can be non-coding RNA, directly functional or be the intermediate protein biosynthesis, template for the synthesis of a protein. The transmission of genes to an organism's offspring, is the basis of the inheritance of phenotypic traits from one generation to the next. These genes make up different DNA sequences, together called a genotype, that is specific to every given individual, within the gene pool of the population (biology), population of a given species. The genotype, along with environmental and developmental factors, ultimately determines the phenotype ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, reading frames are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an open reading frame (ORF) may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Whole Genome Sequencing
Whole genome sequencing (WGS), also known as full genome sequencing or just genome sequencing, is the process of determining the entirety of the DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondrial DNA, mitochondria and, for plants, in the chloroplast. Whole genome sequencing has largely been used as a research tool, but was being introduced to clinics in 2014. In the future of personalized medicine, whole genome sequence data may be an important tool to guide therapeutic intervention. The tool of DNA sequencing, gene sequencing at Single-nucleotide polymorphism, SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology, and hence may lay the foundation for predicting disease susceptibility and drug response. Whole genome sequencing should not be confused with DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulator Gene
In genetics, a regulator gene, regulator, or regulatory gene is a gene involved in controlling the expression of one or more other genes. Regulatory sequences, which encode regulatory genes, are often at the five prime end (5') to the start site of transcription of the gene they regulate. In addition, these sequences can also be found at the three prime end (3') to the transcription start site. In both cases, whether the regulatory sequence occurs before (5') or after (3') the gene it regulates, the sequence is often many kilobases away from the transcription start site. A regulator gene may encode a protein, or it may work at the level of RNA, as in the case of genes encoding microRNAs. An example of a regulator gene is a gene that codes for a repressor protein that inhibits the activity of an operator (a gene which binds repressor proteins thus inhibiting the translation of RNA to protein via RNA polymerase). In prokaryotes, regulator genes often code for repressor pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repetitive DNA
Repeated sequences (also known as repetitive elements, repeating units or repeats) are short or long patterns that occur in multiple copies throughout the genome. In many organisms, a significant fraction of the genomic DNA is repetitive, with over two-thirds of the sequence consisting of repetitive elements in humans. Some of these repeated sequences are necessary for maintaining important genome structures such as telomeres or centromeres. Repeated sequences are categorized into different classes depending on features such as structure, length, location, origin, and mode of multiplication. The disposition of repetitive elements throughout the genome can consist either in directly adjacent arrays called tandem repeats or in repeats dispersed throughout the genome called interspersed repeats. Tandem repeats and interspersed repeats are further categorized into subclasses based on the length of the repeated sequence and/or the mode of multiplication. While some repeated DNA seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Noncoding DNA
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules (e.g. transfer RNA, microRNA, piRNA, ribosomal RNA, and regulatory RNAs). Other functional regions of the non-coding DNA fraction include regulatory sequences that control gene expression; scaffold attachment regions; origins of DNA replication; centromeres; and telomeres. Some non-coding regions appear to be mostly nonfunctional, such as introns, pseudogenes, intergenic DNA, and fragments of transposons and viruses. Regions that are completely nonfunctional are called junk DNA. Fraction of non-coding genomic DNA In bacteria, the coding regions typically take up 88% of the genome. The remaining 12% does not encode proteins, but much of it still has biological function through genes where the RNA transcript is functional (non-coding genes) and regulatory sequences, which means that almost ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed; however, according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin. Constitutive heterochromatin can affect the genes near itself (e.g. position-effect variegation). It is usually repetitive and forms structural functions such as centromeres or telomeres, in addition to acting as an attractor for other gene-expression or repression signals. Facultativ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ENCODE
The Encyclopedia of DNA Elements (ENCODE) is a public research project which aims "to build a comprehensive parts list of functional elements in the human genome." ENCODE also supports further biomedical research by "generating community resources of genomics data, software, tools and methods for genomics data analysis, and products resulting from data analyses and interpretations." The current phase of ENCODE (2016-2019) is adding depth to its resources by growing the number of cell types, data types, assays and now includes support for examination of the mouse genome. History ENCODE was launched by the US National Human Genome Research Institute (NHGRI) in September 2003. Intended as a follow-up to the Human Genome Project, the ENCODE project aims to identify all functional elements in the human genome. The project involves a worldwide consortium of research groups, and data generated from this project can be accessed through public databases. The initial release of ENCODE ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (biology)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription (genetics), transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, Upstream and downstream (DNA), upstream on the DNA (towards the Directionality (molecular biology)#5′-end, 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Overview For transcription to take place, the enzyme that synthesizes RNA, known as RNA polymerase, must attach to the DNA near a gene. Promoters contain specific DNA sequences such as response elements that provide a secure initial binding site for RNA polymerase and for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome. History The term ''exon'' is a shortening of the phrase ''expressed region'' and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron... must be replaced by that of a transcription unit containing regions which will be lost from the mature messengerwhich I suggest we call introns (for intragenic regions)alternating with regions which will be expressedexons." This definition was originally made for protein-coding transcripts that are spliced before b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
De Novo Gene Birth
''De novo'' gene birth is the process by which new genes evolve from non-coding DNA. '' De novo'' genes represent a subset of novel genes, and may be protein-coding or instead act as RNA genes. The processes that govern ''de novo'' gene birth are not well understood, although several models exist that describe possible mechanisms by which ''de novo'' gene birth may occur. Although ''de novo'' gene birth may have occurred at any point in an organism's evolutionary history, ancient ''de novo'' gene birth events are difficult to detect. Most studies of ''de novo'' genes to date have thus focused on young genes, typically taxonomically restricted genes (TRGs) that are present in a single species or lineage, including so-called orphan genes, defined as genes that lack any identifiable homolog. It is important to note, however, that not all orphan genes arise ''de novo'', and instead may emerge through fairly well characterized mechanisms such as gene duplication (including retropo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
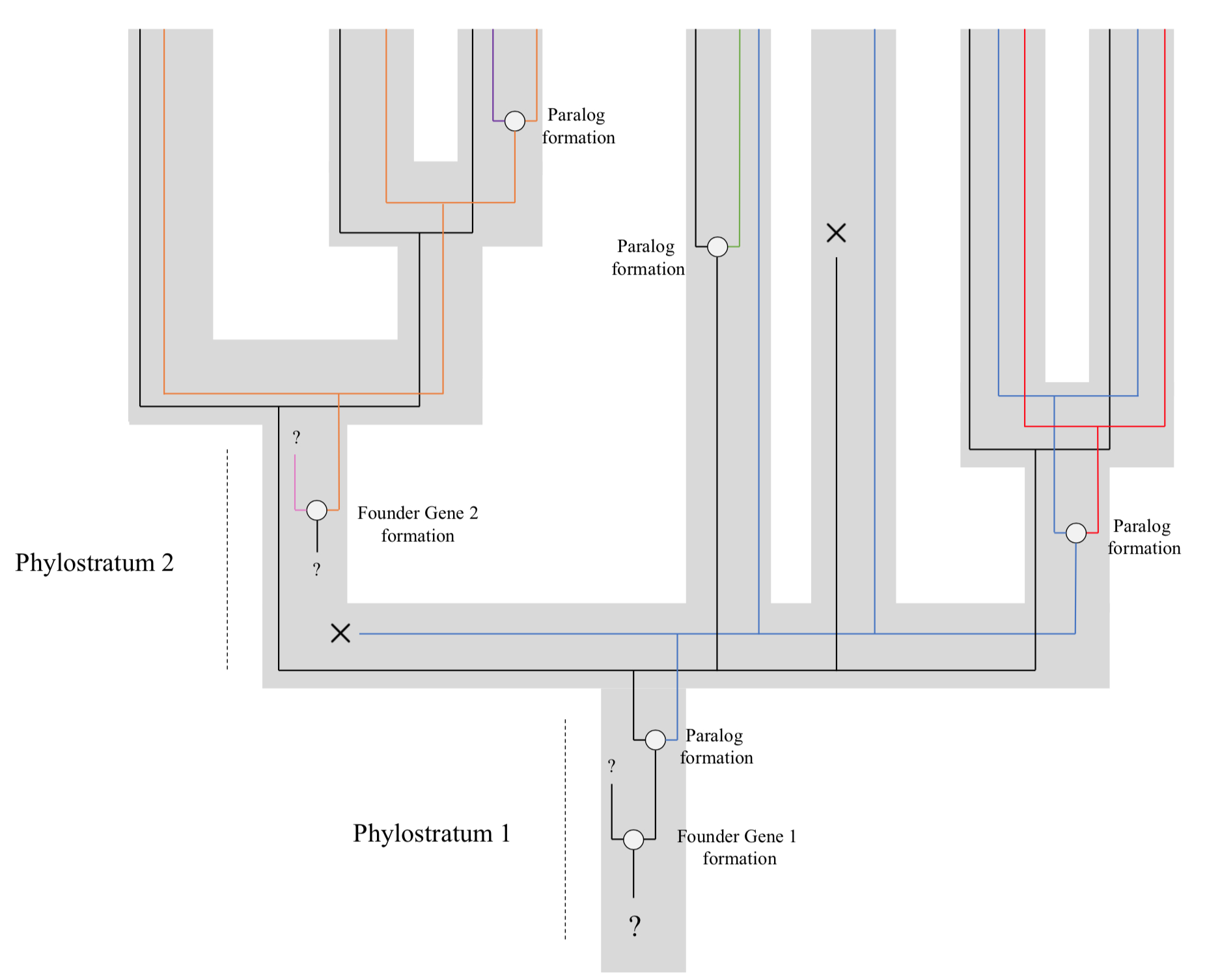
Genomic Phylostratigraphy
Genomic phylostratigraphy is a novel genetic statistical method developed in order to date the origin of specific genes by looking at its homologs across species. It was first developed by Ruđer Bošković Institute in Zagreb, Croatia. The system links genes to their founder gene, allowing us to then determine their age. This could help us better understand many evolutionary processes such as patterns of gene birth throughout evolution, or the relationship between the age of a transcriptome throughout embryonic development. Bioinformatic tools likGenErahave been developed to calculate relative gene ages based on genomic phylostratigraphy. Method This technique relies on the assumption that the diversity of the genome is not only due to gene duplications but also to continuous frequent de novo gene births. These genes (called "founder genes") would form from non-genic DNA sequences, as well as from changes in reading frame (or other ways of arising from within existing genes), ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |