|
Gracilibacteria
Gracilibacteria is a bacterial candidate phylum formerly known as GN02, BD1-5, or SN-2. It is part of the Candidate Phyla Radiation and the Patescibacteria group. The first representative of the Gracilibacteria phylum was reported in 1999 after being recovered from a deep-sea sediment sample. The representative 16S rRNA sequence was referred to as "BD1-5" (sample BD1, sequence 5) and while it was noted that it displayed low sequence identity to any known 16S rRNA gene, it was not proposed as a new phylum at this time. In 2006, representatives of Gracilibacteria were recovered from a hypersaline microbial mat from Guerrero Negro, Baja California Sur, Mexico and proposed as a new phylum "GN02". The BD1-5/GN02 phylum was renamed "Gracilibacteria" in 2013. The first Gracilibacteria genome was recovered from an acetate-amended aquifer (Rifle, CO, USA) using culture-independent, genome-resolved metagenomic techniques in 2012. Genomic analyses suggest that members of the Gracilibacte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacterial Phyla
Bacterial phyla constitute the major lineages of the domain ''Bacteria''. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla. It has been estimated that ~1,300 bacterial phyla exist. As of May 2020, 41 bacterial phyla are formally accepted by the LPSN, 89 bacterial phyla are recognized on thSilva database dozens more have been proposed, and hundreds likely remain to be discovered. As of 2017, approximately 72% of widely recognized bacterial phyla were candidate phyla (i.e. have no cultured representatives). There are no fixed rules to the nomenclature of bacterial phyla. It was proposed that the suffix "-bacteria" be used for phyla. List of bacterial phyla The following is a list of bacterial phyla that have been proposed. Supergroups Despite the unclear ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
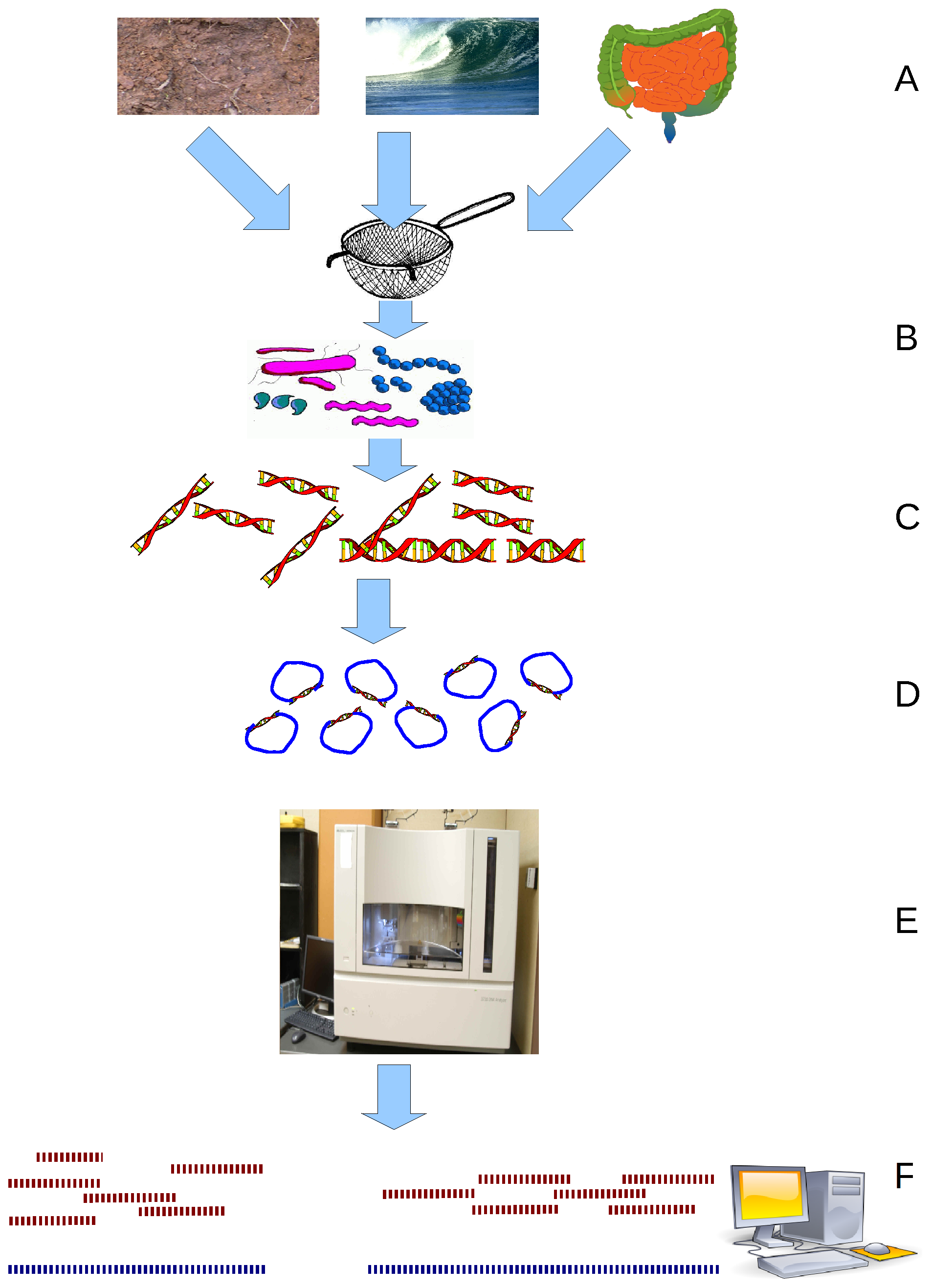
CPR Group
The candidate phyla radiation (also referred to as CPR group) is a large evolutionary radiation of bacterial lineages whose members are mostly uncultivated and only known from metagenomics and single cell sequencing. They have been described as nanobacteria (not to be confused with non-living nanoparticles of the same name) or ultra-small bacteria due to their reduced size (nanometric) compared to other bacteria. Originally (circa 2016), it has been suggested that CPR represents over 15% of all bacterial diversity and may consist of more than 70 different phyla. However, the Genome Taxonomy Database (2018) based on relative evolutionary divergence found that CPR represents a single phylum, with earlier figures inflated by the rapid evolution of ribosomal proteins. CPR lineages are generally characterized as having small genomes and lacking several biosynthetic pathways and ribosomal proteins. This has led to the speculation that they are likely obligate symbionts. Earlie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Candidate Phyla Radiation
The candidate phyla radiation (also referred to as CPR group) is a large evolutionary radiation of bacterial lineages whose members are mostly uncultivated and only known from metagenomics and single cell sequencing. They have been described as nanobacteria (not to be confused with non-living nanoparticles of the same name) or ultra-small bacteria due to their reduced size (nanometric) compared to other bacteria. Originally (circa 2016), it has been suggested that CPR represents over 15% of all bacterial diversity and may consist of more than 70 different phyla. However, the Genome Taxonomy Database (2018) based on relative evolutionary divergence found that CPR represents a single phylum, with earlier figures inflated by the rapid evolution of ribosomal proteins. CPR lineages are generally characterized as having small genomes and lacking several biosynthetic pathways and ribosomal proteins. This has led to the speculation that they are likely obligate symbionts. Earlier work ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metagenomics
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Symbiosis
Symbiosis (from Greek , , "living together", from , , "together", and , bíōsis, "living") is any type of a close and long-term biological interaction between two different biological organisms, be it mutualistic, commensalistic, or parasitic. The organisms, each termed a symbiont, must be of different species. In 1879, Heinrich Anton de Bary defined it as "the living together of unlike organisms". The term was subject to a century-long debate about whether it should specifically denote mutualism, as in lichens. Biologists have now abandoned that restriction. Symbiosis can be obligatory, which means that one or more of the symbionts depend on each other for survival, or facultative (optional), when they can generally live independently. Symbiosis is also classified by physical attachment. When symbionts form a single body it is called conjunctive symbiosis, while all other arrangements are called disjunctive symbiosis."symbiosis." Dorland's Illustrated Medical Dictionary ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endosymbiont
An ''endosymbiont'' or ''endobiont'' is any organism that lives within the body or cells of another organism most often, though not always, in a mutualistic relationship. (The term endosymbiosis is from the Greek: ἔνδον ''endon'' "within", σύν ''syn'' "together" and βίωσις ''biosis'' "living".) Examples are nitrogen-fixing bacteria (called rhizobia), which live in the root nodules of legumes, single-cell algae inside reef-building corals and bacterial endosymbionts that provide essential nutrients to insects. There are two types of symbiont transmissions. In horizontal transmission, each new generation acquires free living symbionts from the environment. An example is the nitrogen-fixing bacteria in certain plant roots. Vertical transmission takes place when the symbiont is transferred directly from parent to offspring. It is also possible for both to be involved in a mixed-mode transmission, where symbionts are transferred vertically for some generation bef ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Code
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries. The codons specify which amino acid will be added next during protein biosynthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. The vast majority of genes are encoded with a single scheme (see the RNA codon table). That scheme is often referred to as the canonical or standard genetic code, or simply ''the'' genetic code, though variant codes (such as in mitochondria) exist. History Effo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycine
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid ( carbamic acid is unstable), with the chemical formula NH2‐ CH2‐ COOH. Glycine is one of the proteinogenic amino acids. It is encoded by all the codons starting with GG (GGU, GGC, GGA, GGG). Glycine is integral to the formation of alpha-helices in secondary protein structure due to its compact form. For the same reason, it is the most abundant amino acid in collagen triple-helices. Glycine is also an inhibitory neurotransmitter – interference with its release within the spinal cord (such as during a '' Clostridium tetani'' infection) can cause spastic paralysis due to uninhibited muscle contraction. It is the only achiral proteinogenic amino acid. It can fit into hydrophilic or hydrophobic environments, due to its minimal side chain of only one hydrogen atom. History and etymology Glycine was discovered in 1820 by the French ch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid ''residues'' form the second-largest component ( water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stop Codon
In molecular biology (specifically protein biosynthesis), a stop codon (or termination codon) is a codon ( nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain. While start codons need nearby sequences or initiation factors to start translation, a stop codon alone is sufficient to initiate termination. Properties Standard codons In the standard genetic code, there are three different termination codons: Alternative stop codons There are variations on the standard genetic code, and alternative stop codons have been found in the mitochondrial genomes of vertebrates, '' Scenedesmus obliquus'', and ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |