|
GSEA
Gene set enrichment analysis (GSEA) (also called functional enrichment analysis or pathway enrichment analysis) is a method to identify classes of genes or proteins that are over-represented in a large set of genes or proteins, and may have an association with different phenotypes (e.g. different organism growth patterns or diseases). The method uses statistical approaches to identify significantly enriched or depleted groups of genes. Transcriptomics technologies and proteomics results often identify thousands of genes, which are used for the analysis. Researchers performing high-throughput experiments that yield sets of genes (for example, genes that are differentially expressed under different conditions) often want to retrieve a functional profile of that gene set, in order to better understand the underlying biological processes. This can be done by comparing the input gene set to each of the bins (terms) in the gene ontology – a statistical test can be performed for eac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome. RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/ SNPs and changes in gene expression over time, or differences in gene expression in different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, small RNA, such as miRNA, tRNA, and ribosomal profiling. RNA-Seq can also be used to determine exon/intron boundaries and verify or amend previously annotated 5' and 3' gene boundaries. Recent advances in RNA-Seq include single cell sequencing, bulk RNA sequencing, 3' mRNA-sequencing, in situ sequencing of fixed tissue, and native RNA molecule sequencin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Odds Ratio
An odds ratio (OR) is a statistic that quantifies the strength of the association between two events, A and B. The odds ratio is defined as the ratio of the odds of event A taking place in the presence of B, and the odds of A in the absence of B. Due to symmetry, odds ratio reciprocally calculates the ratio of the odds of B occurring in the presence of A, and the odds of B in the absence of A. Two events are independent if and only if the OR equals 1, i.e., the odds of one event are the same in either the presence or absence of the other event. If the OR is greater than 1, then A and B are associated (correlated) in the sense that, compared to the absence of B, the presence of B raises the odds of A, and symmetrically the presence of A raises the odds of B. Conversely, if the OR is less than 1, then A and B are negatively correlated, and the presence of one event reduces the odds of the other event occurring. Note that the odds ratio is symmetric in the two events, and no causa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P-value
In null-hypothesis significance testing, the ''p''-value is the probability of obtaining test results at least as extreme as the result actually observed, under the assumption that the null hypothesis is correct. A very small ''p''-value means that such an extreme observed outcome would be very unlikely ''under the null hypothesis''. Even though reporting ''p''-values of statistical tests is common practice in academic publications of many quantitative fields, misinterpretation and misuse of p-values is widespread and has been a major topic in mathematics and metascience. In 2016, the American Statistical Association (ASA) made a formal statement that "''p''-values do not measure the probability that the studied hypothesis is true, or the probability that the data were produced by random chance alone" and that "a ''p''-value, or statistical significance, does not measure the size of an effect or the importance of a result" or "evidence regarding a model or hypothesis". That ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Probability Density Function
In probability theory, a probability density function (PDF), density function, or density of an absolutely continuous random variable, is a Function (mathematics), function whose value at any given sample (or point) in the sample space (the set of possible values taken by the random variable) can be interpreted as providing a ''relative likelihood'' that the value of the random variable would be equal to that sample. Probability density is the probability per unit length, in other words, while the ''absolute likelihood'' for a continuous random variable to take on any particular value is 0 (since there is an infinite set of possible values to begin with), the value of the PDF at two different samples can be used to infer, in any particular draw of the random variable, how much more likely it is that the random variable would be close to one sample compared to the other sample. More precisely, the PDF is used to specify the probability of the random variable falling ''within ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cincinnati Children's Hospital Medical Center
Cincinnati Children's Hospital Medical Center (CCHMC) is an Teaching hospital, academic pediatric acute care children's hospital located in the Avondale, Cincinnati, Avondale neighborhood of Cincinnati, Ohio. The hospital has more than 670 registered beds and is affiliated with the University of Cincinnati Health. The hospital provides comprehensive pediatric specialties and subspecialties to pediatric patients aged 0–21 throughout Appalachian Ohio, southern Ohio and northern Kentucky, as well as patients from around the United States and the world. Cincinnati Children's Hospital Medical Center also treats adults, including adults with congenital heart disease and young adults with blood disease or cancer. Cincinnati Children's Hospital Medical Center also features a Level 1 Trauma Center, Level 1 Pediatric Trauma Center, 1 of 4 in the state. Cincinnati Children's is home to a large neonatology department that oversees newborn nurseries at local hospitals around Ohio. The hospit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
InterMine
InterMine is an open source data warehouse system, licensed under the LGPL 2.1. InterMine is used to create databases of biological data accessed by sophisticated web query tools. InterMine can be used to create databases from a single data set or can integrate multiple sources of data. Support is provided for several common biological formats and there is a framework for adding other data. InterMine includes a user-friendly web interface that works 'out of the box' and can be easily customised. InterMine makes it easy to integrate multiple data sources into a single data warehouse. It has a core data model based on the sequence ontology and supports several biological data formats, allowing sysadmins to configure which organisms or data files are required. It is easy to extend the data model and integrate other data, with a web service API, clients in seven different languages, and an XML format to help import custom data. As an active open source project, InterMine maintains de ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Omics
Omics is the collective characterization and quantification of entire sets of biological molecules and the investigation of how they translate into the structure, function, and dynamics of an organism or group of organisms. The branches of science known informally as ''omics'' are various disciplines in biology whose names end in the suffix ''wikt:-omics, -omics'', such as genomics, proteomics, metabolomics, metagenomics, phenomics and transcriptomics. The related suffix -ome is used to address the objects of study of such fields, such as the genome, proteome or metabolome respectively. The suffix ''-ome'' as used in molecular biology refers to a ''totality'' of some sort; it is an example of a "neo-suffix" formed by abstraction from various Greek terms in , a sequence that does not form an identifiable suffix in Greek. Functional genomics aims at identifying the functions of as many genes as possible of a given organism. It combines different -omics techniques such as transcr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ChIP-seq
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with Massively parallel signature sequencing, massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global binding sites precisely for any protein of interest. Previously, ChIP-on-chip was the most common technique utilized to study these protein–DNA relations. Uses ChIP-seq is primarily used to determine how transcription factors and other chromatin-associated proteins influence phenotype-affecting mechanisms. Determining how proteins interact with DNA to regulate gene expression is essential for fully understanding many biological processes and disease states. This epigenetic information is complementary to genotype and expression analysis. ChIP-seq technology is currently seen primarily as an alternative to ChIP-chip which requires a protein microarray, hybridization ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
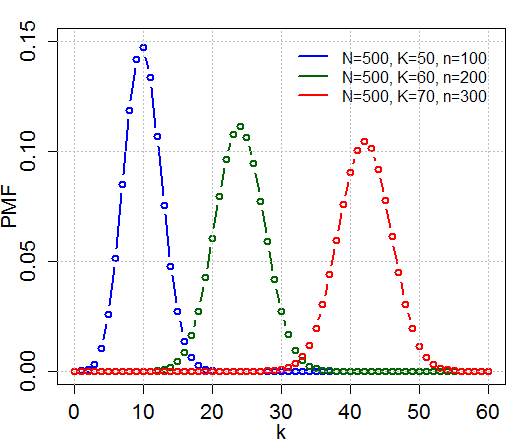
Hypergeometric Test
In probability theory and statistics, the hypergeometric distribution is a Probability distribution#Discrete probability distribution, discrete probability distribution that describes the probability of k successes (random draws for which the object drawn has a specified feature) in n draws, ''without'' replacement, from a finite Statistical population, population of size N that contains exactly K objects with that feature, wherein each draw is either a success or a failure. In contrast, the binomial distribution describes the probability of k successes in n draws ''with'' replacement. Definitions Probability mass function The following conditions characterize the hypergeometric distribution: * The result of each draw (the elements of the population being sampled) can be classified into one of Binary variable, two mutually exclusive categories (e.g. Pass/Fail or Employed/Unemployed). * The probability of a success changes on each draw, as each draw decreases the population ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Ontology
The Gene Ontology (GO) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to: 1) maintain and develop its controlled vocabulary of gene and gene product attributes; 2) annotate genes and gene products, and assimilate and disseminate annotation data; and 3) provide tools for easy access to all aspects of the data provided by the project, and to enable functional interpretation of experimental data using the GO, for example via enrichment analysis. GO is part of a larger classification effort, the Open Biomedical Ontologies, being one of the Initial Candidate Members of the OBO Foundry. Whereas gene nomenclature focuses on gene and gene products, the Gene Ontology focuses on the function of the genes and gene products. The GO also extends the effort by using a markup language to make the data (not only of the genes and their products but also of curated attributes) machine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metascape
Metascape is a free gene annotation and analysis resource that helps biologists make sense of one or multiple gene lists. Metascape provides automated meta-analysis tools to understand either common or unique pathways and protein networks within a group of orthogonal target-discovery studies. History In the "OMICs" age, it is important to gain biological insights into a list of genes. Although a number of bioinformatics sources exist for this purpose, such as DAVID, they are not all free, easy to use, and well maintained. To analyze multiple lists of genes originated from orthogonal but complementary "OMICs" studies, tools often require computational skills that are beyond the reach of many biologists. According to the Metascape blog, a team of scientists self-organized to address this challenge. The team includes core members Yingyao Zhou, Bin Zhou, Lars Pache, Max Chang, Christopher Benner, and Sumit Chanda, as well aother contributorsover the time. Metascape was first re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |