|
FlexAID
FlexAID is a molecular docking software that can use small molecules and peptides as ligands and proteins and nucleic acids as docking targets. As the name suggests, FlexAID supports full ligand flexibility as well side-chain flexibility of the target. It does using a soft scoring function based on the complementarity of the two surfaces (ligand and target). FlexAID has been shown to outperform existing widely used software such as AutoDock Vina and FlexX in the prediction of binding poses. This is particularly true in cases where target flexibility is crucial, such as is likely to be the case when using homology models. The source code is available on GitHub under Apache License. Graphical user interface A PyMOL plugin for FlexAID, NRGsuite, has also been developed by the original authors. See also *Docking (molecular) *Virtual screening Virtual screening (VS) is a computational technique used in drug discovery to search libraries of small molecules in order to identify tho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Protein-ligand Docking Software
The number of notable protein-ligand docking programs currently available is high and has been steadily increasing over the last decades. The following list presents an overview of the most common notable programs, listed alphabetically, with indication of the corresponding year of publication, involved organisation or institution, short description, availability of a webservice and the license. This table is comprehensive but not complete. References External links * {{Chemistry software Structural bioinformatics software Molecular modelling software Computational chemistry software protein-ligand docking ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein–ligand Docking
Protein–ligand docking is a molecular modelling technique. The goal of protein–ligand docking is to predict the position and orientation of a ligand (a small molecule) when it is bound to a protein receptor or enzyme. Pharmaceutical research employs docking techniques for a variety of purposes, most notably in the virtual screening of large databases of available chemicals in order to select likely drug candidates. There has been rapid development in computational ability to determine protein structure with programs such as AlphaFold, and the demand for the corresponding protein-ligand docking predictions is driving implementation of software that can find accurate models. Once the protein folding can be predicted accurately along with how the ligands of various structures will bind to the protein, the ability for drug development to progress at a much faster rate becomes possible. History Computer-aided drug design (CADD) was introduced in the 1980s in order to screen for nove ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
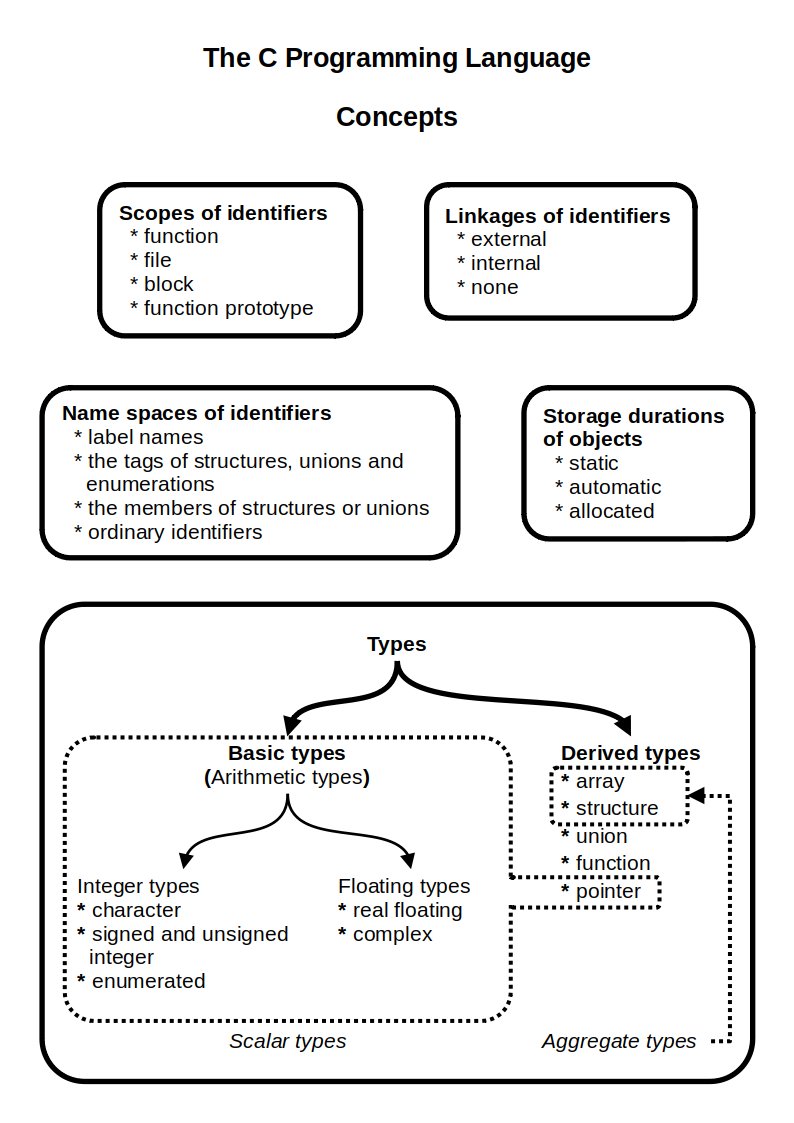
C (programming Language)
C (''pronounced'' '' – like the letter c'') is a general-purpose programming language. It was created in the 1970s by Dennis Ritchie and remains very widely used and influential. By design, C's features cleanly reflect the capabilities of the targeted Central processing unit, CPUs. It has found lasting use in operating systems code (especially in Kernel (operating system), kernels), device drivers, and protocol stacks, but its use in application software has been decreasing. C is commonly used on computer architectures that range from the largest supercomputers to the smallest microcontrollers and embedded systems. A successor to the programming language B (programming language), B, C was originally developed at Bell Labs by Ritchie between 1972 and 1973 to construct utilities running on Unix. It was applied to re-implementing the kernel of the Unix operating system. During the 1980s, C gradually gained popularity. It has become one of the most widely used programming langu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apache License
The Apache License is a permissive free software license written by the Apache Software Foundation (ASF). It allows users to use the software for any purpose, to distribute it, to modify it, and to distribute modified versions of the software under the terms of the license, without concern for royalties. The ASF and its projects release their software products under the Apache License. The license is also used by many non-ASF projects. History Beginning in 1995, the Apache Group (later the Apache Software Foundation) released successive versions of the Apache HTTP Server. Its initial license was essentially the same as the original 4-clause BSD license, with only the names of the organizations changed, and with an additional clause forbidding derivative works from bearing the Apache name. In July 1999, the Berkeley Software Distribution accepted the argument put to it by the Free Software Foundation and retired their ''advertising clause'' (clause 3) to form the new 3-clau ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Docking (molecular)
In the field of molecular modeling, docking is a method which predicts the preferred orientation of one molecule to a second when a ligand and a target are bound to each other to form a stable complex. Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity between two molecules using, for example, scoring functions. The associations between biologically relevant molecules such as proteins, peptides, nucleic acids, carbohydrates, and lipids play a central role in signal transduction. Furthermore, the relative orientation of the two interacting partners may affect the type of signal produced (e.g., agonism vs antagonism). Therefore, docking is useful for predicting both the strength and type of signal produced. Molecular docking is one of the most frequently used methods in structure-based drug design, due to its ability to predict the binding-conformation of small molecule ligands to the appropriate target bind ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
AutoDock Vina
AutoDock is a molecular modeling simulation software. It is especially effective for protein-ligand docking. AutoDock 4 is available under the GNU General Public License. AutoDock is one of the most cited docking software applications in the research community. It is used by the FightAIDS@Home and OpenPandemics - COVID-19 projects run at World Community Grid, to search for antivirals against HIV/AIDS and COVID-19. In February 2007, a search of the ISI Citation Index showed more than 1,100 publications had been cited using the primary AutoDock method papers. As of 2009, this number surpassed 1,200. AutoDock Vina is a successor of AutoDock, significantly improved in terms of accuracy and performance. It is available under the Apache license. Both AutoDock and Vina are currently maintained by Scripps Research, specifically the Center for Computational Structural Biology (CCSB) led by Dr. Arthur J. Olson AutoDock is widely used and played a role in the development of the first cl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GitHub
GitHub () is a Proprietary software, proprietary developer platform that allows developers to create, store, manage, and share their code. It uses Git to provide distributed version control and GitHub itself provides access control, bug tracking system, bug tracking, software feature requests, task management, continuous integration, and wikis for every project. Headquartered in California, GitHub, Inc. has been a subsidiary of Microsoft since 2018. It is commonly used to host open source software development projects. GitHub reported having over 100 million developers and more than 420 million Repository (version control), repositories, including at least 28 million public repositories. It is the world's largest source code host Over five billion developer contributions were made to more than 500 million open source projects in 2024. About Founding The development of the GitHub platform began on October 19, 2005. The site was launched in April 2008 by Tom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PyMOL
PyMOL is a source-available molecular visualization system created by Warren Lyford DeLano. It was commercialized initially by DeLano Scientific LLC, which was a private software company dedicated to creating useful tools that become universally accessible to scientific and educational communities. It is currently commercialized by Schrödinger, Inc. As the original software license was a permissive licence, they were able to remove it; new versions are no longer released under the Python license, but under a custom license (granting broad use, redistribution, and modification rights, but assigning copyright to any version to Schrödinger, LLC.), and some of the source code is no longer released. PyMOL can produce high-quality 3D images of small molecules and biological macromolecules, such as proteins. PyMOL is widely used. PyMOL is one of the few mostly open-source model visualization tools available for use in structural biology. The ''Py'' part of the software's name refe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virtual Screening
Virtual screening (VS) is a computational technique used in drug discovery to search libraries of small molecules in order to identify those structures which are most likely to bind to a drug target, typically a protein receptor (biochemistry), receptor or enzyme. Virtual screening has been defined as "automatically evaluating very large libraries of compounds" using computer programs. As this definition suggests, VS has largely been a numbers game focusing on how the enormous chemical space of over 1060 conceivable compounds can be filtered to a manageable number that can be synthesized, purchased, and tested. Although searching the entire chemical universe may be a theoretically interesting problem, more practical VS scenarios focus on designing and optimizing targeted combinatorial libraries and enriching libraries of available compounds from in-house compound repositories or vendor offerings. As the accuracy of the method has increased, virtual screening has become an integra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Modelling Software
A molecule is a group of two or more atoms that are held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions that satisfy this criterion. In quantum physics, organic chemistry, and biochemistry, the distinction from ions is dropped and ''molecule'' is often used when referring to polyatomic ions. A molecule may be homonuclear, that is, it consists of atoms of one chemical element, e.g. two atoms in the oxygen molecule (O2); or it may be heteronuclear, a chemical compound composed of more than one element, e.g. water (two hydrogen atoms and one oxygen atom; H2O). In the kinetic theory of gases, the term ''molecule'' is often used for any gaseous particle regardless of its composition. This relaxes the requirement that a molecule contains two or more atoms, since the noble gases are individual atoms. Atoms and complexes connected by non-covalent interactions, such as hydrogen bonds or ionic bonds, are typically ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Modelling
Molecular modelling encompasses all methods, theoretical and computational, used to model or mimic the behaviour of molecules. The methods are used in the fields of computational chemistry, drug design, computational biology and materials science to study molecular systems ranging from small chemical systems to large biological molecules and material assemblies. The simplest calculations can be performed by hand, but inevitably computers are required to perform molecular modelling of any reasonably sized system. The common feature of molecular modelling methods is the atomistic level description of the molecular systems. This may include treating atoms as the smallest individual unit (a molecular mechanics approach), or explicitly modelling protons and neutrons with its quarks, anti-quarks and gluons and electrons with its photons (a quantum chemistry approach). Molecular mechanics Molecular mechanics is one aspect of molecular modelling, as it involves the use of classical mec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Free And Open-source Software
Free and open-source software (FOSS) is software available under a license that grants users the right to use, modify, and distribute the software modified or not to everyone free of charge. FOSS is an inclusive umbrella term encompassing free software and open-source software. The rights guaranteed by FOSS originate from the "Four Essential Freedoms" of '' The Free Software Definition'' and the criteria of '' The Open Source Definition''. All FOSS can have publicly available source code, but not all source-available software is FOSS. FOSS is the opposite of proprietary software, which is licensed restrictively or has undisclosed source code. The historical precursor to FOSS was the hobbyist and academic public domain software ecosystem of the 1960s to 1980s. Free and open-source operating systems such as Linux distributions and descendants of BSD are widely used, powering millions of servers, desktops, smartphones, and other devices. Free-software licenses and open-so ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |