|
Fastq
FASTQ format is a text-based format for storing both a biological sequence (usually nucleotide sequence) and its corresponding quality scores. Both the sequence letter and quality score are each encoded with a single ASCII character for brevity. It was originally developed at the Wellcome Trust Sanger Institute to bundle a FASTA formatted sequence and its quality data, but has recently become the ''de facto'' standard for storing the output of high-throughput sequencing instruments such as the Illumina Genome Analyzer. Format A FASTQ file has four line-separated fields per sequence: * Field 1 begins with a '@' character and is followed by a sequence identifier and an ''optional'' description (like a FASTA title line). * Field 2 is the raw sequence letters. * Field 3 begins with a '+' character and is ''optionally'' followed by the same sequence identifier (and any description) again. * Field 4 encodes the quality values for the sequence in Field 2, and must contain the same nu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FASTA Format
In bioinformatics and biochemistry, the FASTA format is a text-based format for representing either nucleotide sequences or amino acid (protein) sequences, in which nucleotides or amino acids are represented using single-letter codes. The format also allows for sequence names and comments to precede the sequences. The format originates from the FASTA software package, but has now become a near universal standard in the field of bioinformatics. The simplicity of FASTA format makes it easy to manipulate and parse sequences using text-processing tools and scripting languages like the R programming language, Python, Ruby, Haskell, and Perl. Original format & overview The original FASTA/ Pearson format is described in the documentation for the FASTA suite of programs. It can be downloaded with any free distribution of FASTA (see fasta20.doc, fastaVN.doc or fastaVN.me—where VN is the Version Number). In the original format, a sequence was represented as a series of lines, each o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phred Quality Score
A Phred quality score is a measure of the quality of the identification of the nucleobases generated by automated DNA sequencing. It was originally developed for the computer program Phred to help in the automation of DNA sequencing in the Human Genome Project. Phred quality scores are assigned to each nucleotide base call in automated sequencer traces. The FASTQ format encodes phred scores as ASCII characters alongside the read sequences. Phred quality scores have become widely accepted to characterize the quality of DNA sequences, and can be used to compare the efficacy of different sequencing methods. Perhaps the most important use of Phred quality scores is the automatic determination of accurate, quality-based consensus sequences. Definition Phred quality scores Q are logarithmically related to the base-calling error probabilities P and defined as Q = -10 \ \log_ P. This relation can be also be written as P = 10^. For example, if Phred assigns a quality score of 30 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Burrows–Wheeler Transform
The Burrows–Wheeler transform (BWT, also called block-sorting compression) rearranges a character string into runs of similar characters. This is useful for compression, since it tends to be easy to compress a string that has runs of repeated characters by techniques such as move-to-front transform and run-length encoding. More importantly, the transformation is ''reversible'', without needing to store any additional data except the position of the first original character. The BWT is thus a "free" method of improving the efficiency of text compression algorithms, costing only some extra computation. The Burrows–Wheeler transform is an algorithm used to prepare data for use with data compression techniques such as bzip2. It was invented by Michael Burrows and David Wheeler in 1994 while Burrows was working at DEC Systems Research Center in Palo Alto, California. It is based on a previously unpublished transformation discovered by Wheeler in 1983. The algorithm can be imp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coverage (genetics)
Coverage (or depth) in DNA sequencing is the number of unique reads that include a given nucleotide in the reconstructed sequence. Deep sequencing refers to the general concept of aiming for high number of unique reads of each region of a sequence. Rationale Even though the sequencing accuracy for each individual nucleotide is very high, the very large number of nucleotides in the genome means that if an individual genome is only sequenced once, there will be a significant number of sequencing errors. Furthermore, many positions in a genome contain rare single-nucleotide polymorphisms (SNPs). Hence to distinguish between sequencing errors and true SNPs, it is necessary to increase the sequencing accuracy even further by sequencing individual genomes a large number of times. Ultra-deep sequencing The term "ultra-deep" can sometimes also refer to higher coverage (>100-fold), which allows for detection of sequence variants in mixed populations. In the extreme, error-corrected sequ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAM (file Format)
Sequence Alignment Map (SAM) is a text-based format originally for storing biological sequences aligned to a reference sequence developed by Heng Li and Bob Handsaker ''et al''. It was developed when the 1000 Genomes Project wanted to move away from the MAQ mapper format and decided to design a new format. The overall TAB-delimited flavour of the format came from an earlier format inspired by BLAT’s PSL. The name of SAM came from Gabor Marth from University of Utah, who originally had a format under the same name but with a different syntax more similar to a BLAST output. It is widely used for storing data, such as nucleotide sequences, generated by next generation sequencing technologies, and the standard has been broadened to include unmapped sequences. The format supports short and long reads (up to 128 Mbp) produced by different sequencing platforms and is used to hold mapped data within the Genome Analysis Toolkit (GATK) and across the Broad Institute, the Wellcome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
De Bruijn Graph
In graph theory, an -dimensional De Bruijn graph of symbols is a directed graph representing overlaps between sequences of symbols. It has vertices, consisting of all possible sequences of the given symbols; the same symbol may appear multiple times in a sequence. For a set of symbols the set of vertices is: :V=S^n=\. If one of the vertices can be expressed as another vertex by shifting all its symbols by one place to the left and adding a new symbol at the end of this vertex, then the latter has a directed edge to the former vertex. Thus the set of arcs (that is, directed edges) is :E=\. Although De Bruijn graphs are named after Nicolaas Govert de Bruijn, they were discovered independently by both De Bruijn and I. J. Good. Much earlier, Camille Flye Sainte-Marie implicitly used their properties. Properties * If , then the condition for any two vertices forming an edge holds vacuously, and hence all the vertices are connected, forming a total of edges. * Each vertex h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
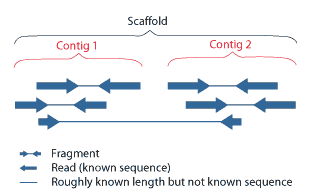
Contig
A contig (from ''contiguous'') is a set of overlapping DNA segments that together represent a consensus region of DNA.Gregory, S. ''Contig Assembly''. Encyclopedia of Life Sciences, 2005. In bottom-up sequencing projects, a contig refers to overlapping sequence data ( reads); in top-down sequencing projects, contig refers to the overlapping clones that form a physical map of the genome that is used to guide sequencing and assembly.Dear, P. H. ''Genome Mapping''. Encyclopedia of Life Sciences, 2005. . Contigs can thus refer both to overlapping DNA sequences and to overlapping physical segments (fragments) contained in clones depending on the context. Original definition of contig In 1980, Staden wrote: ''In order to make it easier to talk about our data gained by the shotgun method of sequencing we have invented the word "contig". A contig is a set of gel readings that are related to one another by overlap of their sequences. All gel readings belong to one and only one cont ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Wellcome Trust Sanger Institute
The Wellcome Sanger Institute, previously known as The Sanger Centre and Wellcome Trust Sanger Institute, is a non-profit British genomics and genetics research institute, primarily funded by the Wellcome Trust. It is located on the Wellcome Genome Campus by the village of Hinxton, outside Cambridge. It shares this location with the European Bioinformatics Institute. It was established in 1992 and named after double Nobel Laureate Frederick Sanger. It was conceived as a large scale DNA sequencing centre to participate in the Human Genome Project, and went on to make the largest single contribution to the gold standard sequence of the human genome. From its inception the institute established and has maintained a policy of data sharing, and does much of its research in collaboration. Since 2000, the institute expanded its mission to understand "the role of genetics in health and disease". The institute now employs around 900 people and engages in five main areas of research: Canc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
File Extension
A filename extension, file name extension or file extension is a suffix to the name of a computer file (e.g., .txt, .docx, .md). The extension indicates a characteristic of the file contents or its intended use. A filename extension is typically delimited from the rest of the filename with a full stop (period), but in some systems it is separated with spaces. Other extension formats include dashes and/or underscores on early versions of Linux and some versions of IBM AIX. Some file systems implement filename extensions as a feature of the file system itself and may limit the length and format of the extension, while others treat filename extensions as part of the filename without special distinction. Usage Filename extensions may be considered a type of metadata. They are commonly used to imply information about the way data might be stored in the file. The exact definition, giving the criteria for deciding what part of the file name is its extension, belongs to the rules of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hierarchical Data Format
Hierarchical Data Format (HDF) is a set of file formats (HDF4, HDF5) designed to store and organize large amounts of data. Originally developed at the U.S. National Center for Supercomputing Applications, it is supported by The HDF Group, a non-profit corporation whose mission is to ensure continued development of HDF5 technologies and the continued accessibility of data stored in HDF. In keeping with this goal, the HDF libraries and associated tools are available under a liberal, BSD-like license for general use. HDF is supported by many commercial and non-commercial software platforms and programming languages. The freely available HDF distribution consists of the library, command-line utilities, test suite source, Java interface, and the Java-based HDF Viewer (HDFView). The current version, HDF5, differs significantly in design and API from the major legacy version HDF4. Early history The quest for a portable scientific data format, originally dubbed AEHOO (All Encompassing ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |