|
Fas Receptor
The Fas receptor, also known as Fas, FasR, apoptosis antigen 1 (APO-1 or APT), cluster of differentiation 95 (CD95) or tumor necrosis factor receptor superfamily member 6 (TNFRSF6), is a protein that in humans is encoded by the ''FAS'' gene. Fas was first identified using a monoclonal antibody generated by immunizing mice with the FS-7 cell line. Thus, the name Fas is derived from ''F''S-7-''a''ssociated ''s''urface antigen. The Fas receptor is a death receptor on the surface of cells that leads to programmed cell death (apoptosis) if it binds its ligand, Fas ligand (FasL). It is one of two apoptosis pathways, the other being the mitochondrial pathway. Gene FAS receptor gene is located on the long arm of chromosome 10 (10q24.1) in humans and on chromosome 19 in mice. The gene lies on the plus ( Watson strand) and is 25,255 bases in length organized into nine protein encoding exons. Similar sequences related by evolution ( orthologs) are found in most mammals. Protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cluster Of Differentiation
The cluster of differentiation (also known as cluster of designation or classification determinant and often abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules providing targets for immunophenotyping of cells. In terms of physiology, CD molecules can act in numerous ways, often acting as receptors or ligands important to the cell. A signal cascade is usually initiated, altering the behavior of the cell (see cell signaling). Some CD proteins do not play a role in cell signaling, but have other functions, such as cell adhesion. CD for humans is numbered up to 371 (). Nomenclature The CD nomenclature was proposed and established in the 1st International Workshop and Conference on Human Leukocyte Differentiation Antigens (HLDA), held in Paris in 1982. This system was intended for the classification of the many monoclonal antibodies (mAbs) generated by different laboratories around the world against epitopes on the surface mo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alternative Splicing
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene may be included within or excluded from the final RNA product of the gene. This means the exons are joined in different combinations, leading to different splice variants. In the case of protein-coding genes, the proteins translated from these splice variants may contain differences in their amino acid sequence and in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most of the observed splice variants ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tumor Suppressor
A tumor suppressor gene (TSG), or anti-oncogene, is a gene that regulates a cell (biology), cell during cell division and replication. If the cell grows uncontrollably, it will result in cancer. When a tumor suppressor gene is mutated, it results in a loss or reduction in its function. In combination with other genetic mutations, this could allow the cell to grow abnormally. The Loss-of-function mutation, loss of function for these genes may be even more significant in the development of human cancers, compared to the activation of oncogenes. TSGs can be grouped into the following categories: caretaker genes, gatekeeper genes, and more recently landscaper genes. Caretaker genes ensure stability of the genome via DNA repair and subsequently when mutated allow mutations to accumulate. Meanwhile, gatekeeper genes directly regulate cell growth by either inhibiting cell cycle progression or inducing apoptosis. Lastly, landscaper genes regulate growth by contributing to the surrounding e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
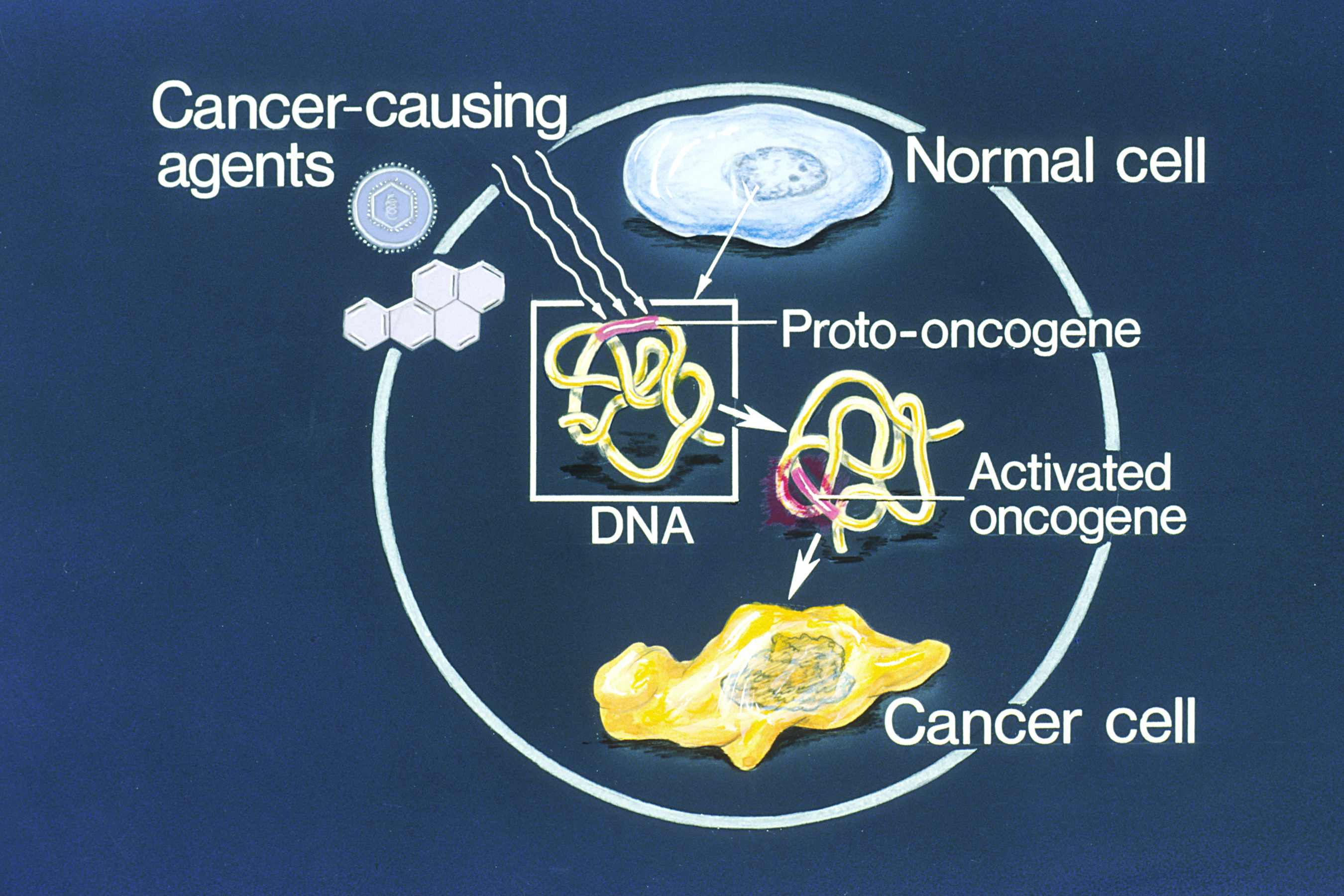
Oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.Kimball's Biology Pages. "Oncogenes" Free full text Most normal cells undergo a preprogrammed rapid cell death () if critical functions are altered and then malfunction. Activated oncogenes can cause those cells designated for apoptosis to survive and proliferate instead. Most oncogenes began as proto-oncogenes: normal genes involved in cell growth and proliferation or inhibition of apoptosis. If, through mutation, normal genes promoting cellular growth are up-regulated (gain-of-function mutation), they predispose the cel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteolytic Cleavage
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Protein degradation is a major regulatory mechanism of gene expression and contributes substantially to shaping mammalian proteomes. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion. Proteolysis in organisms serves many purposes; for example, digestive enzymes break down proteins in food to provide amino acids for the organism, while proteolytic processing of a polypeptide chain after its synthesis may be necessary for the production of an active protein. It is also important in the regulation of some physiological and cellular processes including apoptosis, as well as preventing the accumulation of unwanted or misfolded proteins in cells. Consequently, abnormality in the regulation of proteolysis can cause diseases. Proteolysis can also ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caspase-8
Caspase-8 is a caspase protein, encoded by the ''CASP8'' gene. It most likely acts upon caspase-3. ''CASP8'' orthologs have been identified in numerous mammals for which complete genome data are available. These unique orthologs are also present in birds. Function The ''CASP8'' gene encodes a member of the cysteine-aspartic acid protease (caspase) family. Sequential activation of caspases plays a central role in the execution-phase of cell apoptosis. Caspases exist as inactive proenzymes composed of a prodomain, a large protease Protein subunit, subunit, and a small protease subunit. Activation of caspases requires proteolytic processing at conserved internal aspartic residues to generate a heterodimeric enzyme consisting of the large and small subunits. This protein is involved in the programmed cell death induced by Fas receptor, Fas and various apoptotic stimuli. The N-terminal FADD-like death effector domain of this protein suggests that it may interact with Fas-interacting ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Death Effector Domain
The death-effector domain (DED) is a protein interaction domain found only in eukaryotes that regulates a variety of cellular signalling pathways. The DED domain is found in inactive procaspases (cysteine proteases) and proteins that regulate caspase activation in the apoptosis cascade such as FAS-associating death domain-containing protein (FADD). FADD recruits procaspase 8 and procaspase 10 into a death induced signaling complex (DISC). This recruitment is mediated by a homotypic interaction between the procaspase DED and a second DED that is death effector domain in an Signal transducing adaptor protein, adaptor protein that is directly associated with activated TNF receptors. Complex formation allows proteolytic activation of procaspase into the active caspase form which results in the initiation of apoptosis (cell death). Structurally the DED domain are a subclass of protein motif known as the death fold and contains 6 alpha helices, that closely resemble the structure of the D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FADD
FAS-associated death domain protein, also called MORT1, is encoded by the ''FADD'' gene on the 11q13.3 region of chromosome 11 in humans. FADD is an Signal transducing adaptor protein, adaptor protein that bridges members of the Tumor necrosis factor receptor, tumor necrosis factor receptor superfamily, such as the FasR, Fas-receptor, to caspase 8, procaspases 8 and caspase 10, 10 to form the death-inducing signaling complex (DISC) during apoptosis. As well as its most well known role in apoptosis, FADD has also been seen to play a role in other processes including proliferation, cell cycle regulation and development. Structure FADD is a 23 kDa protein, made up of 208 amino acids. It contains two main domains: a C terminal death domain (DD) and an N terminal death effector domain (DED). Each domain, although sharing very little sequence similarity, are structurally similar to one another, with each consisting of 6 α helices. The DD of FADD binds to receptors such as the Fas r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Transducing Adaptor Protein
Signal transducing adaptor proteins (STAPs) are proteins that are accessory to main proteins in a signal transduction pathway. Adaptor proteins contain a variety of protein-binding modules that link protein-binding partners together and facilitate the creation of larger signaling complexes. These proteins tend to lack any intrinsic enzymatic activity themselves, instead mediating specific protein–protein interactions that drive the formation of Multiprotein complex, protein complexes. Examples of adaptor proteins include MYD88, Grb2 and SHC1. Signaling components Much of the specificity of signal transduction depends on the recruitment of several signalling components such as protein kinases and G-protein GTPases into short-lived active complexes in response to an activating signal such as a growth factor binding to its receptor (biochemistry), receptor. Domains Adaptor proteins usually contain several domains within their structure (e.g., SH2 domain, Src homology 2 (SH2) and S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endosome
Endosomes are a collection of intracellular sorting organelles in eukaryotic cells. They are parts of the endocytic membrane transport pathway originating from the trans Golgi network. Molecules or ligands internalized from the plasma membrane can follow this pathway all the way to lysosomes for degradation or can be recycled back to the cell membrane in the endocytic cycle. Molecules are also transported to endosomes from the trans Golgi network and either continue to lysosomes or recycle back to the Golgi apparatus. Endosomes can be classified as early, sorting, or late depending on their stage post internalization. Endosomes represent a major sorting compartment of the endomembrane system in cells. Function Endosomes provide an environment for material to be sorted before it reaches the degradative lysosome. For example, low-density lipoprotein (LDL) is taken into the cell by binding to the LDL receptor at the cell surface. Upon reaching early endosomes, the LDL dis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Death-inducing Signaling Complex
The death-inducing signaling complex (DISC) is a multiprotein complex formed by members of the death receptor family of apoptosis-inducing cellular receptors. A typical example is FasR, which forms the DISC upon trimerization as a result of its ligand ( FasL) binding. The DISC is composed of the death receptor, FADD, and caspase 8. It transduces a downstream signal cascade resulting in apoptosis. Description The Fas ligands, or cytotoxicity-dependent APO-1-associated proteins, physically associate with APO-1 (also known as the Fas receptor, or CD95), a tumor necrosis factor containing a functional death domain. This association leads to the formation of the DISC, thereby inducing apoptosis. The entire process is initiated when the cell registers the presence of CD95L, the cognate ligand for APO-1. Upon binding, the CAP proteins and procaspase-8 (composed of FLICE, MACH, and Mch5) bind to CD95 through death domain and death effector domain interactions. Procaspase-8 activa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cysteine
Cysteine (; symbol Cys or C) is a semiessential proteinogenic amino acid with the chemical formula, formula . The thiol side chain in cysteine enables the formation of Disulfide, disulfide bonds, and often participates in enzymatic reactions as a nucleophile. Cysteine is chiral, but both D and L-cysteine are found in nature. LCysteine is a protein monomer in all biota, and D-cysteine acts as a signaling molecule in mammalian nervous systems. Cysteine is named after its discovery in urine, which comes from the urinary bladder or cyst, from Ancient Greek, Greek κύστις ''kýstis'', "bladder". The thiol is susceptible to oxidation to give the disulfide bond, disulfide derivative cystine, which serves an important structural role in many proteins. In this case, the symbol Cyx is sometimes used. The deprotonated form can generally be described by the symbol Cym as well. When used as a food additive, cysteine has the E number E920. Cysteine is Genetic code, encoded by the codo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |