|
FAM83H
FAM83H is a gene in humans that encodes a protein known as FAM83H (uncharacterized protein FAM83H). FAM83H is targeted for the nucleus and it predicted to play a role in the structural development and calcification of tooth enamel. Gene Location FAM83H is located on the long arm of chromosome 8 (8q24.3), starting at 143723933 and ending at 143738030. The FAM83H gene spans 14097 base pairs and is orientated on the—strand. The coding region is made up of 5,604 base pairs and 5 exons. Expression FAM83H is ubiquitously expressed throughout the human body at relatively low levels. Transcript Variants In humans, there is only one known major product of the FAM83H gene. Homology Paralogs There are no paralogs of FAM83H Orthologs Below is a table of a variety of orthologs of the human FAM83H. The table include closely, intermediately and distantly related orthologs. Orthologs of the human protein FAM83H are listed above in descending order or date of divergence and the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome 8
Chromosome 8 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 8 spans about 145 million base pairs (the building material of DNA) and represents between 4.5 and 5.0% of the total DNA in cells. About 8% of its genes are involved in brain development and function, and about 16% are involved in cancer. A unique feature of 8p is a region of about 15 megabases that appears to have a high mutation rate. This region shows a significant divergence between human and chimpanzee, suggesting that its high mutation rates have contributed to the evolution of the human brain. __TOC__ Genes Number of genes The following are some of the gene count estimates of human chromosome 8. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phospholipase
A phospholipase is an enzyme that hydrolyzes phospholipids into fatty acids and other lipophilic substances. Acids trigger the release of bound calcium from cellular stores and the consequent increase in free cytosolic Ca2+, an essential step in calcium signaling to regulate intracellular processes. There are four major classes, termed A, B, C, and D, which are distinguished by the type of reaction which they catalyze: *Phospholipase A ** Phospholipase A1 – cleaves the ''sn''-1 acyl chain (where ''sn'' refers to stereospecific numbering). **Phospholipase A2 – cleaves the ''sn''-2 acyl chain, releasing arachidonic acid. *Phospholipase B – cleaves both ''sn''-1 and ''sn''-2 acyl chains; this enzyme is also known as a lysophospholipase. *Phospholipase C – cleaves before the phosphate, releasing diacylglycerol and a phosphate-containing head group. PLCs play a central role in signal transduction, releasing the second messenger inositol triphosphate. * Phospholipase D – ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MMP20
Matrix metalloproteinase-20 (MMP-20) also known as enamel metalloproteinase or enamelysin is an enzyme that in humans is encoded by the ''MMP20'' gene. Function Proteins of the matrix metalloproteinase ( MMP) family are involved in the breakdown of extracellular matrix in normal physiological processes, such as embryonic development, reproduction, and tissue remodeling, as well as in disease processes, such as arthritis and metastasis. Most MMP's are secreted as inactive proproteins which are activated when cleaved by extracellular proteinases. MMP-20, also known as enamelysin, appears to be the only MMP that is tooth-specific and it is expressed by cells of different developmental origin (i.e. epithelial ameloblasts and mesenchymal odontoblasts). Clinical significance The human MMP-20 gene contains 10 exons and is part of a cluster of matrix metalloproteinase genes that localize to human chromosome 11q22.3. A mutation in this gene, which alters the normal splice pattern and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WDR72
WD repeat-containing protein 72 is a protein that in humans is encoded by the ''WDR72'' gene. WDR72 contains 7 WD40 repeats, which are predicted to form the blades of a 7 beta propeller structure. Clinical significance Mutations in this gene cause autosomal-recessive hypomaturation amelogenesis imperfecta Amelogenesis imperfecta (AI) is a congenital disorder which presents with a rare abnormal formation of the enamel or external layer of the crown of teeth, unrelated to any systemic or generalized conditions. Enamel is composed mostly of mineral, .... References Further reading * * * * * * * * {{gene-15-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Interacting Proteins
Interaction is action that occurs between two or more objects, with broad use in philosophy and the sciences. It may refer to: Science * Interaction hypothesis, a theory of second language acquisition * Interaction (statistics) * Interactions of actors theory, created by cybernetician Gordon Pask * Fundamental interaction, in physics * Human–computer interaction * Social interaction between people Biology * Biological interaction * Cell–cell interaction * Drug interaction * Gene–environment interaction * Protein–protein interaction Chemistry * Aromatic interaction * Cation-pi interaction * Metallophilic interaction Arts and media * ''Interaction'' (album), 1963, by Art Farmer's Quartet * ACM ''Interactions'', a magazine published by the Association for Computing Machinery * "Interactions" (''The Spectacular Spider-Man''), an episode of the animated television series * 63rd World Science Fiction Convention, titled ''Interaction'' See also * * * Interact (disambi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phyre 1
Phyre and Phyre2 (Protein Homology/AnalogY Recognition Engine; pronounced as 'fire') are free web-based services for protein structure prediction. Phyre is among the most popular methods for protein structure prediction having been cited over 1500 times. Like other remote homology recognition techniques (see protein threading), it is able to regularly generate reliable protein models when other widely used methods such as PSI-BLAST cannot. Phyre2 has been designed to ensure a user-friendly interface for users inexpert in protein structure prediction methods. Its development is funded by the Biotechnology and Biological Sciences Research Council. Description The Phyre and Phyre2 servers predict the three-dimensional structure of a protein sequence using the principles and techniques of homology modeling. Because the structure of a protein is more conserved in evolution than its amino acid sequence, a protein sequence of interest (the target) can be modeled with reasonable accuracy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
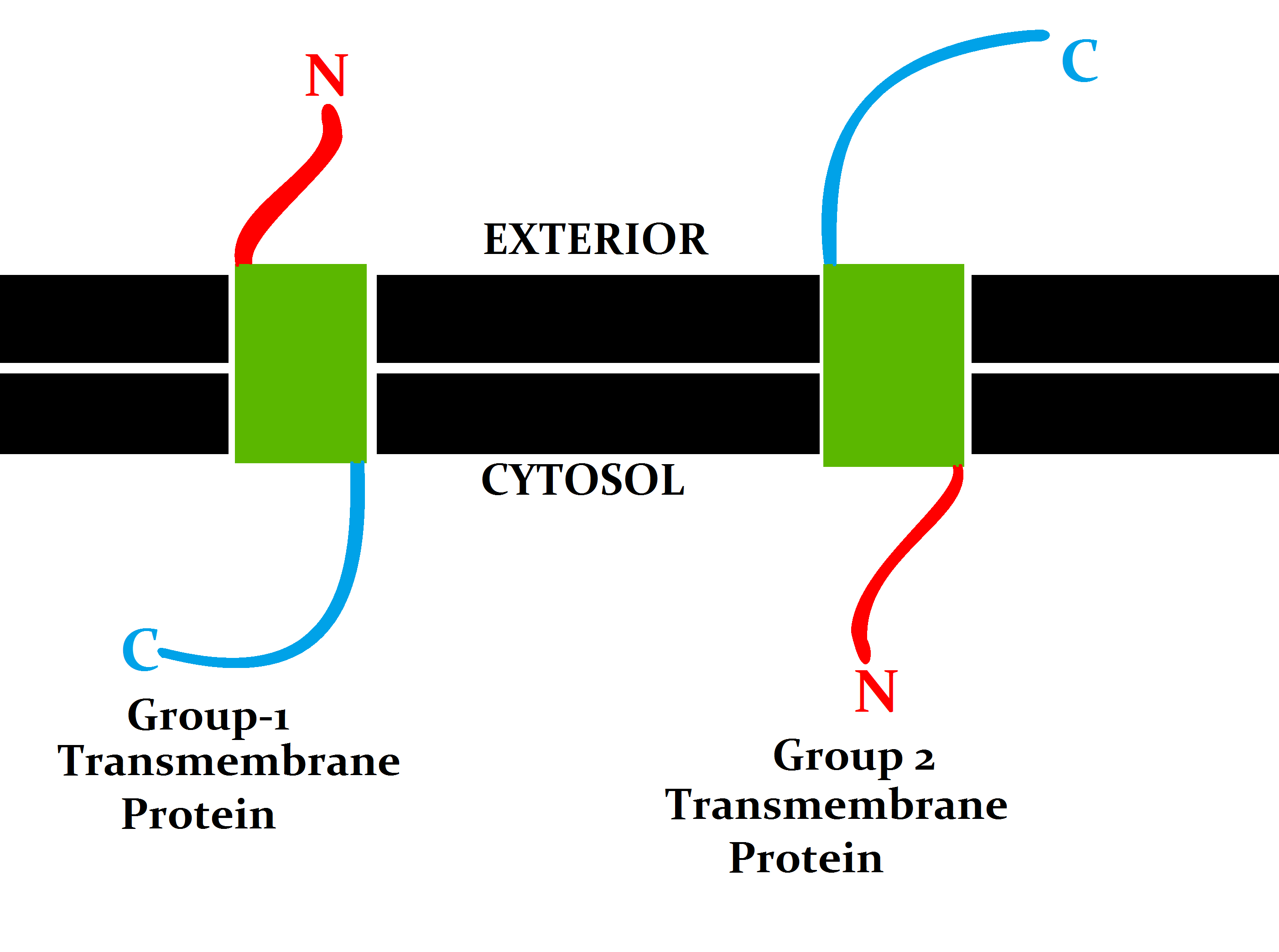
Transmembrane Prediction
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not pass t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helices
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astbu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Types The most common secondary st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Motif Scan 1
Motif may refer to: General concepts * Motif (chess composition), an element of a move in the consideration of its purpose * Motif (folkloristics), a recurring element that creates recognizable patterns in folklore and folk-art traditions * Motif (music), a salient recurring fragment or succession of notes * Motif (narrative), any recurring element in a story that has symbolic significance or the reason behind actions * Motif (textile arts), a recurring element or fragment that, when joined together, creates a larger work * Motif (visual arts), a repeated theme or pattern Biochemistry * Sequence motif, a sequence pattern of nucleotides in a DNA sequence or amino acids in a protein * Short linear motif, a stretch of protein sequence that mediates protein–protein interaction * Structural motif, a pattern in a protein structure formed by the spatial arrangement of amino acids Other uses * ''Motif'' (2019 film), 2019 Malaysian Malay-language crime drama film * ''Motif'' (al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sumoylation
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation (sometimes written sumoylation). SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. SUMO proteins are similar to ubiquitin and are considered members of the ubiquitin-like protein family. SUMOylation is directed by an enzymatic cascade analogous to that involved in ubiquitination. In contrast to ubiquitin, SUMO is not used to tag proteins for degradation. Mature SUMO is produced when the last four amino acids of the C-terminus have been cleaved off to allow formation of an isopeptide bond between the C-terminal glycine residue of SUMO and an acceptor lysine on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |