|
Exonic Splicing Enhancer
In molecular biology, an exonic splicing enhancer (ESE) is a DNA sequence motif consisting of 6 bases within an exon that directs, or enhances, accurate splicing of heterogeneous nuclear RNA ( hnRNA) or pre-mRNA into messenger RNA (mRNA). Introduction Short sequences of DNA are transcribed to RNA; then this RNA is translated to a protein. A gene located in DNA will contain introns and exons. Part of the process of preparing the RNA includes splicing out the introns, sections of RNA that do not code for the protein. The presence of exonic splicing enhancers is essential for proper identification of splice sites by the cellular machinery. Role in splicing SR proteins bind to and promote exon splicing in regions with ESEs, while heterogeneous ribonucleoprotein particles (hnRNPs) bind to and block exon splicing in regions with exonic splicing silencers. Both types of proteins are involved in the assembly and proper functioning of spliceosomes. During RNA splicing, U2 s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactions. Though cells and other microscopic structures had been observed in living organisms as early as the 18th century, a detailed understanding of the mechanisms and interactions governing their behavior did not emerge until the 20th century, when technologies used in physics and chemistry had advanced sufficiently to permit their application in the biological sciences. The term 'molecular biology' was first used in 1945 by the English physicist William Astbury, who described it as an approach focused on discerning the underpinnings of biological phenomena—i.e. uncovering the physical and chemical structures and properties of biological molecules, as well as their interactions with other molecules and how these interactions explain observ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exonic Splicing Silencer
An exonic splicing silencer (ESS) is a short region (usually 4-18 nucleotides) of an exon and is a cis-regulatory element. A set of 103 hexanucleotides known as FAS-hex3 has been shown to be abundant in ESS regions. ESSs inhibit or silence splicing of the pre-mRNA and contribute to constitutive and alternate splicing. To elicit the silencing effect, ESSs recruit proteins that will negatively affect the core splicing machinery. Mechanism of action Exonic splicing silencers work by inhibiting the splicing of pre-mRNA strands or promoting exon skipping. The single stranded pre-mRNA molecules need to have their intronic and exonic regions spliced in order to be translated. ESSs silence splice sites adjacent to them by interfering with the components of the core splicing complex, such as the snRNP's, U1 and U2. This causes proteins that negatively influence splicing to be recruited to the splicing machinery. ESSs have four general roles: * inhibiting exon inclusion * inhibiting in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HNPCC
Hereditary nonpolyposis colorectal cancer (HNPCC) is a hereditary predisposition to colon cancer. HNPCC includes (and was once synonymous with) Lynch syndrome, an autosomal dominant genetic condition that is associated with a high risk of colon cancer, endometrial cancer (second most common), ovary, stomach, small intestine, hepatobiliary tract, upper urinary tract, brain, and skin. The increased risk for these cancers is due to inherited genetic mutations that impair DNA mismatch repair. It is a type of cancer syndrome. Other HNPCC conditions include Lynch-like syndrome, polymerase proofreading-associated polyposis and familial colorectal cancer type X. Signs and symptoms Risk of cancer ''Lifetime risk and mean age at diagnosis for Lynch syndrome–associated cancers'' In addition to the types of cancer found in the chart above, it is understood that Lynch syndrome also contributes to an increased risk of small bowel cancer, pancreatic cancer, ureter/renal pelvis cancer, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MLH1
DNA mismatch repair protein Mlh1 or MutL protein homolog 1 is a protein that in humans is encoded by the ''MLH1'' gene located on chromosome 3. The gene is commonly associated with hereditary nonpolyposis colorectal cancer. Orthologs of human MLH1 have also been studied in other organisms including mouse and the budding yeast ''Saccharomyces cerevisiae''. Function Variants in this gene can cause hereditary nonpolyposis colon cancer (Lynch syndrome). It is a human homolog of the ''E. coli'' DNA mismatch repair gene, mutL, which mediates protein-protein interactions during mismatch recognition, strand discrimination, and strand removal. Defects in MLH1 are associated with the microsatellite instability observed in hereditary nonpolyposis colon cancer. Alternatively spliced transcript variants encoding different isoforms have been described, but their full-length natures have not been determined. Role in DNA mismatch repair MLH1 protein is one component of a system of seven ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
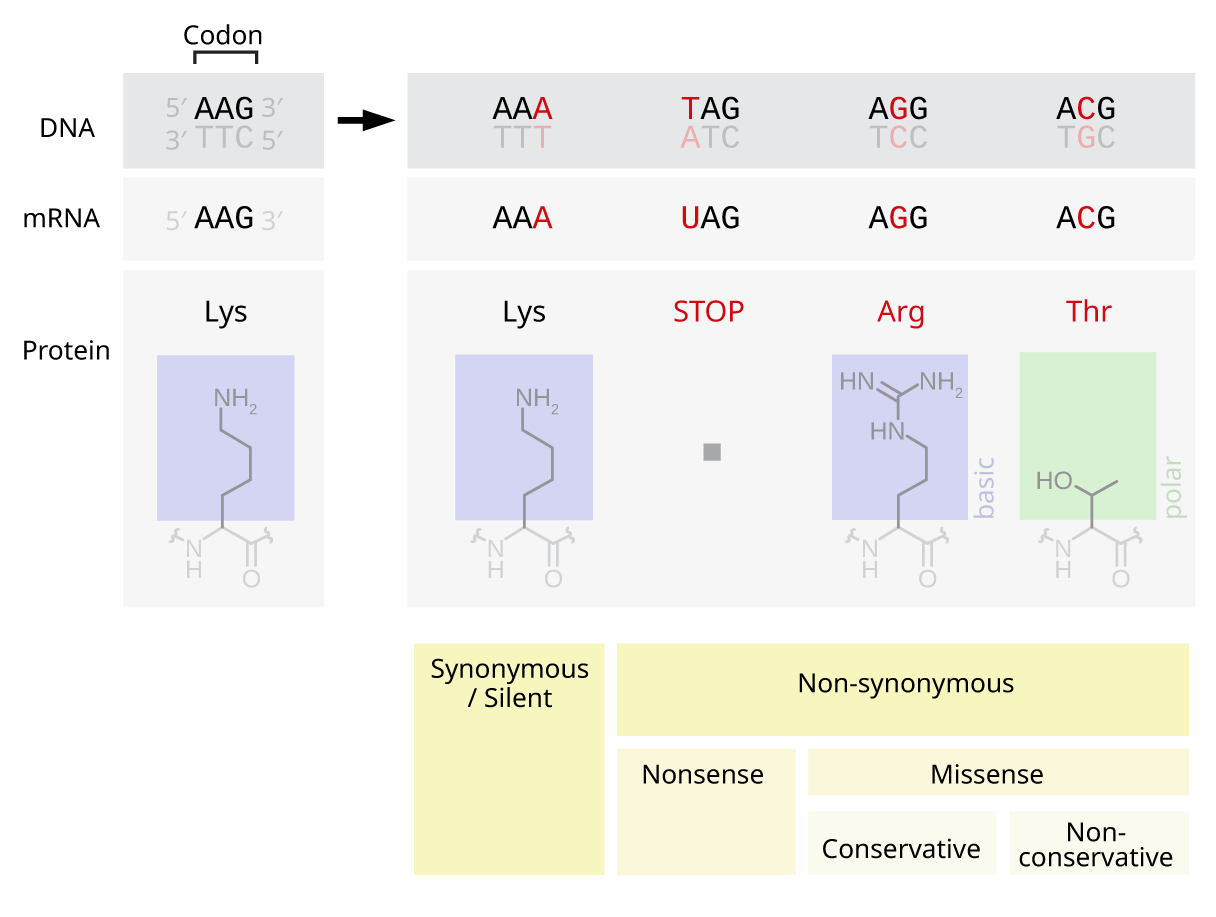
Silent Mutations
Silent mutations, also called synonymous or samesense mutations, are mutations in DNA that do not have an observable effect on the organism's phenotype. The phrase ''silent mutation'' is often used interchangeably with the phrase '' synonymous mutation''; however, synonymous mutations are not always silent, nor vice versa. Synonymous mutations can affect transcription, splicing, mRNA transport, and translation, any of which could alter phenotype, rendering the synonymous mutation non-silent. The substrate specificity of the tRNA to the rare codon can affect the timing of translation, and in turn the co-translational folding of the protein. This is reflected in the codon usage bias that is observed in many species. Mutations that cause the altered codon to produce an amino acid with similar functionality (''e.g.'' a mutation producing leucine instead of isoleucine) are often classified as silent; if the properties of the amino acid are conserved, this mutation does not usually si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
In Vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, and plants, as opposed to a tissue extract or dead organism. Examples of investigations ''in vivo'' include: the pathogenesis of disease by comparing the effects of bacterial infection with the effects of purified bacterial toxins; the development of non-antibiotics, antiviral drugs, and new drugs generally; and new surgical procedures. Consequently, animal testing and clinical trials are major elements of ''in vivo'' research. ''In vivo'' testing is often employed over ''in vitro'' because it is better suited for observing the overall effects of an experiment on a living subject. In drug discovery, for example, verification of efficacy ''in vivo'' is crucial, because ''in vitro'' assays can sometimes yield misleading results with drug c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alternative Splicing
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene may be included within or excluded from the final RNA product of the gene. This means the exons are joined in different combinations, leading to different splice variants. In the case of protein-coding genes, the proteins translated from these splice variants may contain differences in their amino acid sequence and in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most of the observed splice variants ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U2AF2
Splicing factor U2AF 65 kDa subunit is a protein that in humans is encoded by the ''U2AF2'' gene. Function In eukaryotes, the introns in the transcribed pre-mRNA first have to be removed by spliceosome in order to form a mature mRNA. A spliceosome is assembled from small nuclear ribonucleoproteins(snRNP) and small nuclear RNAs(snRNA). And the splicing factor can be divided into snRNP and non snRNP proteins.U2 auxiliary factor ( U2AF), composed of a large and a small subunit, is a non-snRNP protein required for the binding of U2 snRNP to the pre-mRNA branch site. This gene encodes the U2AF large subunit, which contains a sequence-specific RNA-binding region with 3 RNA recognition motifs and an Arg/Ser-rich domain necessary for splicing. The large subunit binds to the polypyrimidine tract of introns early during spliceosome assembly. Multiple alternatively spliced transcript variants have been detected for this gene, but the full-length natures of only two have been determine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U2 Small Nuclear RNA Auxiliary Factor 1
Splicing factor U2AF 35 kDa subunit is a protein that in humans is encoded by the ''U2AF1'' gene. Function This gene belongs to the splicing factor SR family of genes . U2AF1 is a subunit of the U2 Auxiliary Factor complex alongside a larger subunit, U2AF2. U2AF1 is a non-snRNP protein required for the binding of U2 snRNP to the pre-mRNA branch site. This gene encodes a small (~35 kDa) subunit which plays a critical role in RNA splicing by recognizing and binding to AG nucleotides at the 3’ splice site to facilitate spliceosome assembly. Alternatively spliced transcript variants encoding different isoforms have been identified . Somatic mutations in ''U2AF1'' have been found in a range of human cancers, with a distinctive pattern of these mutations at the zinc fingers implicating a functional role under selection. In lung cancers, these mutations affect alternative splicing of several transcripts, including oncogenic ROS1 fusions. U2af1 conditional deletion in mouse hem ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Splicing
RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcription (biology), transcript is transformed into a mature messenger RNA (Messenger RNA, mRNA). It works by removing all the introns (non-coding regions of RNA) and ''splicing'' back together exons (coding regions). For nuclear genes, nuclear-encoded genes, splicing occurs in the cell nucleus, nucleus either during or immediately after Transcription (biology), transcription. For those eukaryotic transcription, eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translation (biology), translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, spli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs ( snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to specific proteins to form a small nuclear ribonucleoprotein complex (snRNP, pronounced "snurps"), which in turn combines with other snRNPs to form a large ribonucleoprotein complex called a spliceosome. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material (equivalent to the introns) from the initial film and sends the cleaned-up version to the director for the final cut. However, sometimes the RNA within the intron acts as a ribozyme, splicing itself without the use of a spliceosome or protein enzymes. History In 1977, work by the Sharp and Roberts labs reve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heterogeneous Ribonucleoprotein Particle
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are complexes of RNA and protein present in the cell nucleus during gene transcription and subsequent post-transcriptional modification of the newly synthesized RNA (pre-mRNA). The presence of the proteins bound to a pre-mRNA molecule serves as a signal that the pre-mRNA is not yet fully processed and therefore not ready for export to the cytoplasm. Since most mature RNA is exported from the nucleus relatively quickly, most RNA-binding protein in the nucleus exist as heterogeneous ribonucleoprotein particles. After splicing has occurred, the proteins remain bound to spliced introns and target them for degradation. hnRNPs are also integral to the 40S subunit of the ribosome and therefore important for the translation of mRNA in the cytoplasm. However, hnRNPs also have their own nuclear localization sequences (NLS) and are therefore found mainly in the nucleus. Though it is known that a few hnRNPs shuttle between the cytoplasm and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |