|
Doctor In A Cell
Doctor in a cell is an advanced biotechnology concept introduced in 1998 by Ehud Shapiro of the Weizmann Institute. It envisions autonomous, programmable molecular devices within the human body performing diagnostic and therapeutic functions. The initial design proposed molecular Turing machines capable of replacing traditional drugs with "smart drugs" composed of molecular computing devices programmed with medical knowledge to analyze their environment and release appropriate treatments. To realize this vision, Shapiro and colleague Kobi Benenson developed molecular implementations at the Weizmann Institute. These included a programmable autonomous automaton with DNA-encoded input and transition rules, and a stochastic automaton that interacts with its environment to release drug molecules in response to cancer markers. In 2009, Shapiro and PhD student Tom Ran created a prototype molecular system capable of simple logical deductions using DNA strands. This system represented the f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ehud Shapiro
Ehud Shapiro (; born 1955) is an Israeli scientist, entrepreneur, artist, and political activist who is Professor of Computer Science and Biology at the Weizmann Institute of Science. With international reputation, he made contributions to many scientific disciplines, laying in each a long-term research agenda by asking a basic question and offering a first step towards answering it, including how to computerize the process of scientific discovery, by providing an algorithmic interpretation to Karl Popper's methodology of conjectures and refutations; how to automate program debugging, by algorithms for fault localization; how to unify parallel, distributed, and systems programming with a high-level logic-based programming language; how to use the metaverse as a foundation for social networking; how to devise molecular computers that can function as smart programmable drugs; how to uncover the human cell lineage tree, via single-cell genomics; how to support digital democracy, by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prostate Cancer
Prostate cancer is the neoplasm, uncontrolled growth of cells in the prostate, a gland in the male reproductive system below the bladder. Abnormal growth of the prostate tissue is usually detected through Screening (medicine), screening tests, typically blood tests that check for prostate-specific antigen (PSA) levels. Those with high levels of PSA in their blood are at increased risk for developing prostate cancer. Diagnosis requires a prostate biopsy, biopsy of the prostate. If cancer is present, the pathologist assigns a Gleason score; a higher score represents a more dangerous tumor. Medical imaging is performed to look for cancer that has spread outside the prostate. Based on the Gleason score, PSA levels, and imaging results, a cancer case is assigned a cancer staging, stage 1 to 4. A higher stage signifies a more advanced, more dangerous disease. Most prostate tumors remain small and cause no health problems. These are managed with active surveillance of prostate cancer, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are approximately 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit the air, soil, water, Hot spring, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria play a vital role in many stages of the nutrient cycle by recycling nutrients and the nitrogen fixation, fixation of nitrogen from the Earth's atmosphere, atmosphere. The nutrient cycle includes the decomposition of cadaver, dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, suc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinians, Augustinian friar working in the 19th century in Brno, was the first to study genetics scientifically. Mendel studied "trait inheritance", patterns in the way traits are handed down from parents to offspring over time. He observed that organisms (pea plants) inherit traits by way of discrete "units of inheritance". This term, still used today, is a somewhat ambiguous definition of what is referred to as a gene. Phenotypic trait, Trait inheritance and Molecular genetics, molecular inheritance mechanisms of genes are still primary principles of genetics in the 21st century, but modern genetics has expanded to study the function and behavior of genes. Gene structure and function, variation, and distribution are studied within the context of the Cell (bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactions. Though cells and other microscopic structures had been observed in living organisms as early as the 18th century, a detailed understanding of the mechanisms and interactions governing their behavior did not emerge until the 20th century, when technologies used in physics and chemistry had advanced sufficiently to permit their application in the biological sciences. The term 'molecular biology' was first used in 1945 by the English physicist William Astbury, who described it as an approach focused on discerning the underpinnings of biological phenomena—i.e. uncovering the physical and chemical structures and properties of biological molecules, as well as their interactions with other molecules and how these interactions explain observ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
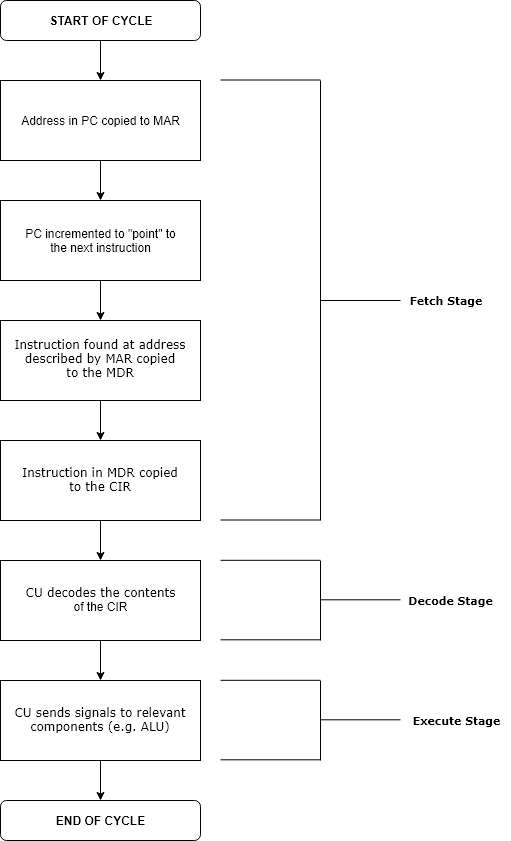
Execution (computing)
Execution in computer and software engineering is the process by which a computer or virtual machine interprets and acts on the instructions of a computer program. Each instruction of a program is a description of a particular action which must be carried out, in order for a specific problem to be solved. Execution involves repeatedly following a " fetch–decode–execute" cycle for each instruction done by the control unit. As the executing machine follows the instructions, specific effects are produced in accordance with the semantics of those instructions. Programs for a computer may be executed in a batch process without human interaction or a user may type commands in an interactive session of an interpreter. In this case, the "commands" are simply program instructions, whose execution is chained together. The term run is used almost synonymously. A related meaning of both "to run" and "to execute" refers to the specific action of a user starting (or ''launching'' or ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
In Vitro
''In vitro'' (meaning ''in glass'', or ''in the glass'') Research, studies are performed with Cell (biology), cells or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in biology and its subdisciplines are traditionally done in labware such as test tubes, flasks, Petri dishes, and microtiter plates. Studies conducted using components of an organism that have been isolated from their usual biological surroundings permit a more detailed or more convenient analysis than can be done with whole organisms; however, results obtained from ''in vitro'' experiments may not fully or accurately predict the effects on a whole organism. In contrast to ''in vitro'' experiments, ''in vivo'' studies are those conducted in living organisms, including humans, known as clinical trials, and whole plants. Definition ''In vitro'' (Latin language, Latin for "in glass"; often not italicized in English usage) studies are conducted ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
In Silico
In biology and other experimental sciences, an ''in silico'' experiment is one performed on a computer or via computer simulation software. The phrase is pseudo-Latin for 'in silicon' (correct ), referring to silicon in computer chips. It was coined in 1987 as an allusion to the Latin phrases , , and , which are commonly used in biology (especially systems biology). The latter phrases refer, respectively, to experiments done in living organisms, outside living organisms, and where they are found in nature. History The earliest known use of the phrase was by Christopher Langton to describe artificial life, in the announcement of a workshop on that subject at the Center for Nonlinear Studies at the Los Alamos National Laboratory in 1987. The expression ''in silico'' was first used to characterize biological experiments carried out entirely in a computer in 1989, in the workshop "Cellular Automata: Theory and Applications" in Los Alamos, New Mexico, by Pedro Miramontes, a mathemat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
High-level Programming Language
A high-level programming language is a programming language with strong Abstraction (computer science), abstraction from the details of the computer. In contrast to low-level programming languages, it may use natural language ''elements'', be easier to use, or may automate (or even hide entirely) significant areas of computing systems (e.g. memory management), making the process of developing a program simpler and more understandable than when using a lower-level language. The amount of abstraction provided defines how "high-level" a programming language is. In the 1960s, a high-level programming language using a compiler was commonly called an ''autocode''. Examples of autocodes are COBOL and Fortran. The first high-level programming language designed for computers was Plankalkül, created by Konrad Zuse. However, it was not implemented in his time, and his original contributions were largely isolated from other developments due to World War II, aside from the language's influe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Compiler
In computing, a compiler is a computer program that Translator (computing), translates computer code written in one programming language (the ''source'' language) into another language (the ''target'' language). The name "compiler" is primarily used for programs that translate source code from a high-level programming language to a lower level language, low-level programming language (e.g. assembly language, object code, or machine code) to create an executable program.Compilers: Principles, Techniques, and Tools by Alfred V. Aho, Ravi Sethi, Jeffrey D. Ullman - Second Edition, 2007 There are many different types of compilers which produce output in different useful forms. A ''cross-compiler'' produces code for a different Central processing unit, CPU or operating system than the one on which the cross-compiler itself runs. A ''bootstrap compiler'' is often a temporary compiler, used for compiling a more permanent or better optimised compiler for a language. Related software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |