|
Dihydropyrimidinase
In enzymology, a dihydropyrimidinase () is an enzyme that catalyzes the chemical reaction :5,6-dihydrouracil + H2O \rightleftharpoons 3-ureidopropanoate Thus, the two substrates of this enzyme are 5,6-dihydrouracil and H2O, whereas its product is 3-ureidopropanoate. This enzyme belongs to the family of hydrolases, those acting on carbon-nitrogen bonds other than peptide bonds, specifically in cyclic amides. The systematic name of this enzyme class is 5,6-dihydropyrimidine amidohydrolase. Other names in common use include hydantoinase, hydropyrimidine hydrase, hydantoin peptidase, pyrimidine hydrase, and D-hydantoinase. This enzyme participates in 3 metabolic pathways In biochemistry, a metabolic pathway is a linked series of chemical reactions occurring within a cell. The reactants, products, and intermediates of an enzymatic reaction are known as metabolites, which are modified by a sequence of chemical ...: pyrimidine metabolism, beta-alanine metabolism, and p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
5,6-dihydrouracil
Dihydrouracil is an intermediate in the catabolism of uracil. It is the base present in the nucleoside dihydrouridine. See also * Dihydrouracil dehydrogenase (NAD+) * Dihydrouracil oxidase * Dihydropyrimidinase In enzymology, a dihydropyrimidinase () is an enzyme that catalyzes the chemical reaction :5,6-dihydrouracil + H2O \rightleftharpoons 3-ureidopropanoate Thus, the two substrates of this enzyme are 5,6-dihydrouracil and H2O, whereas its produ ... References Nucleobases Ureas Pyrimidinediones {{organic-compound-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
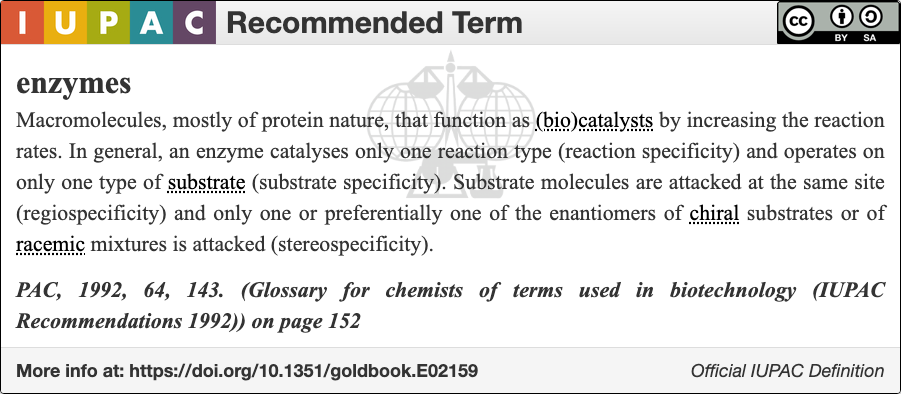
Enzymology
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
Enzymes are listed here by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system: :Oxidoreductases (EC 1) ( Oxidoreductase) * Dehydrogenase * Luciferase * DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) ** Homoserine dehydrogenase ** Aminopropanol oxidoreductase ** Diacetyl reductase ** Glycerol dehydrogenase ** Propanediol-phosphate dehydrogenase ** glycerol-3-phoshitiendopene dehydrogenase (NAD+) ** D-xylulose reductase ** L-xylulose reductase ** Lactate dehydrogenase ** Malate dehydrogenase ** Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) ** Glucose oxidase ** L-gulonolactone oxidase ** Thiamine oxidase ** Xanthine oxidase * EC 1.1.4 (with a disulfide as accep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). This structural data is obtained and deposited by biologists and biochemists worldwide through the use of experimental methodologies such as X-ray crystallography, Nuclear magnetic resonance spectroscopy of proteins, NMR spectroscopy, and, increasingly, cryo-electron microscopy. All submitted data are reviewed by expert Biocuration, biocurators and, once approved, are made freely available on the Internet under the CC0 Public Domain Dedication. Global access to the data is provided by the websites of the wwPDB member organizations (PDBe, PDBj, RCSB PDB, and BMRB). The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the backbone may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure. A number of these structures may bind to each other, forming a quaternary structure. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypeptide chains and amino acid side chains, it was Dorothy Maud Wrinch who incorporated geometry into the prediction of protein structures. Wrinch demon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pantothenate And Coa Biosynthesis
Pantothenic acid (vitamin B5) is a B vitamin and an essential nutrient. All animals need pantothenic acid in order to synthesize coenzyme A (CoA), which is essential for cellular energy production and for the synthesis and degradation of proteins, carbohydrates, and fats. Pantothenic acid is the combination of pantoic acid and β-alanine. Its name comes from the Greek ''pantothen'', meaning "from everywhere", because pantothenic acid, at least in small amounts, is in almost all foods. Deficiency of pantothenic acid is very rare in humans. In dietary supplements and animal feed, the form commonly used is calcium pantothenate, because chemically it is more stable, and hence makes for longer product shelf-life, than sodium pantothenate and free pantothenic acid. Definition Pantothenic acid is a water-soluble vitamin, one of the B vitamins. It is synthesized from the amino acid β-alanine and pantoic acid (see biosynthesis and structure of coenzyme A figures). Unlike vitamin E ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrimidine Metabolism
Pyrimidine biosynthesis occurs both in the body and through organic synthesis. ''De novo'' biosynthesis of pyrimidine ''De Novo'' biosynthesis of a pyrimidine is catalyzed by three gene products CAD, DHODH and UMPS. The first three enzymes of the process are all coded by the same gene in CAD which consists of carbamoyl phosphate synthetase II, aspartate carbamoyltransferase and dihydroorotase. Dihydroorotate dehydrogenase (DHODH) unlike CAD and UMPS is a mono-functional enzyme and is localized in the mitochondria. UMPS is a bifunctional enzyme consisting of orotate phosphoribosyltransferase (OPRT) and orotidine monophosphate decarboxylase (OMPDC). Both, CAD and UMPS are localized around the mitochondria, in the cytosol. In Fungi, a similar protein exists but lacks the dihydroorotase function: another protein catalyzes the second step. In other organisms (Bacteria, Archaea and the other Eukaryota), the first three steps are done by three different enzymes. Pyrimidine c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metabolism
Metabolism (, from ''metabolē'', "change") is the set of life-sustaining chemical reactions in organisms. The three main functions of metabolism are: the conversion of the energy in food to energy available to run cellular processes; the conversion of food to building blocks of proteins, lipids, nucleic acids, and some carbohydrates; and the elimination of metabolic wastes. These enzyme-catalyzed reactions allow organisms to grow and reproduce, maintain their Structures#Biological, structures, and respond to their environments. The word ''metabolism'' can also refer to the sum of all chemical reactions that occur in living organisms, including digestion and the transportation of substances into and between different cells, in which case the above described set of reactions within the cells is called intermediary (or intermediate) metabolism. Metabolic reactions may be categorized as ''catabolic''—the ''breaking down'' of compounds (for example, of glucose to pyruvate by c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrolase
In biochemistry, hydrolases constitute a class of enzymes that commonly function as biochemical catalysts that use water to break a chemical bond: :\ce \quad \xrightarrowtext\quad \ce This typically results in dividing a larger molecule into smaller molecules. Some common examples of hydrolase enzymes are esterases including lipases, phosphatases, glycosidases, peptidases, and nucleosidases. Esterases cleave ester bonds in lipids and phosphatases cleave phosphate groups off molecules. An example of crucial esterase is acetylcholine esterase, which assists in transforming the neuron impulse into the acetate group after the hydrolase breaks the acetylcholine into choline and acetic acid. Acetic acid is an important metabolite in the body and a critical intermediate for other reactions such as glycolysis. Lipases hydrolyze glycerides. Glycosidases cleave sugar molecules off carbohydrates and peptidases hydrolyze peptide bonds. Nucleosidases hydrolyze the bonds of nucleo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
3-ureidopropanoate
3-Ureidopropionic acid, also called N-carbamoyl-beta-alanine, is an intermediate in the metabolism of uracil. It is a urea Urea, also called carbamide (because it is a diamide of carbonic acid), is an organic compound with chemical formula . This amide has two Amine, amino groups (–) joined by a carbonyl functional group (–C(=O)–). It is thus the simplest am ... derivative of beta-alanine. References Propionic acids Ureas {{OrganicAcid-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |