|
David Goodsell
David S. Goodsell, is an associate professor at the Scripps Research Institute and research professor at Rutgers University, New Jersey (joint appointment). He is especially known for his watercolor paintings of cell interiors. Education David Goodsell studied a BSc in biology and chemistry at University of California Irvine. After this, he did a PhD in X-ray crystallography of DNA at the University of California Los Angeles, completed in 1987. Research Since completing his PhD he has worked as a structural biologist at the Scripps Research Institute (with a 2-year period in University of California in 1992-94). His research topics have included the use of structural biology and molecular dynamic simulations to investigate symmetry in protein oligomers, protein-protein interactions and for computer-aided drug design. In particular he is a developer of AutoDock, the most widely-used program used for molecular docking. His main research focus areas are HIV drug res ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Scripps Research Institute
Scripps Research, previously known as The Scripps Research Institute (TSRI), is a nonprofit American medical research facility that focuses on research and education in the biomedical sciences. Headquartered in San Diego, California, the institute has over 170 laboratories employing 2,100 scientists, technicians, graduate students, and administrative and other staff, making it the largest private, non-profit biomedical research organization in the United States and among the largest in the world. The institute holds over 1,100 patents, has produced 11 FDA-approved therapeutics, and has generated over 50 spin-off companies. According to the 2017 Nature Innovation Index, Scripps Research is the #1 most influential research institution in the world. The Scripps Research graduate program is ranked 9th nationally in the biological sciences, 6th for organic chemistry, and 6th for biochemistry. In 2022, their Jupiter, FL campus became a part of the University of Florida. Jupiter-bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multienzyme complexes, in which multiple catalytic domains are found in a single polypeptide chain. Protein complexes are a form of quaternary structure. Proteins in a protein complex are linked by non-covalent protein–protein interactions. These complexes are a cornerstone of many (if not most) biological processes. The cell is seen to be composed of modular supramolecular complexes, each of which performs an independent, discrete biological function. Through proximity, the speed and selectivity of binding interactions between enzymatic complex and substrates can be vastly improved, leading to higher cellular efficiency. Many of the techniques used to enter cells and isolate proteins are inherently disruptive to such large complexes, complicating the task of determining the components of a complex. Examples of protein complexes include th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
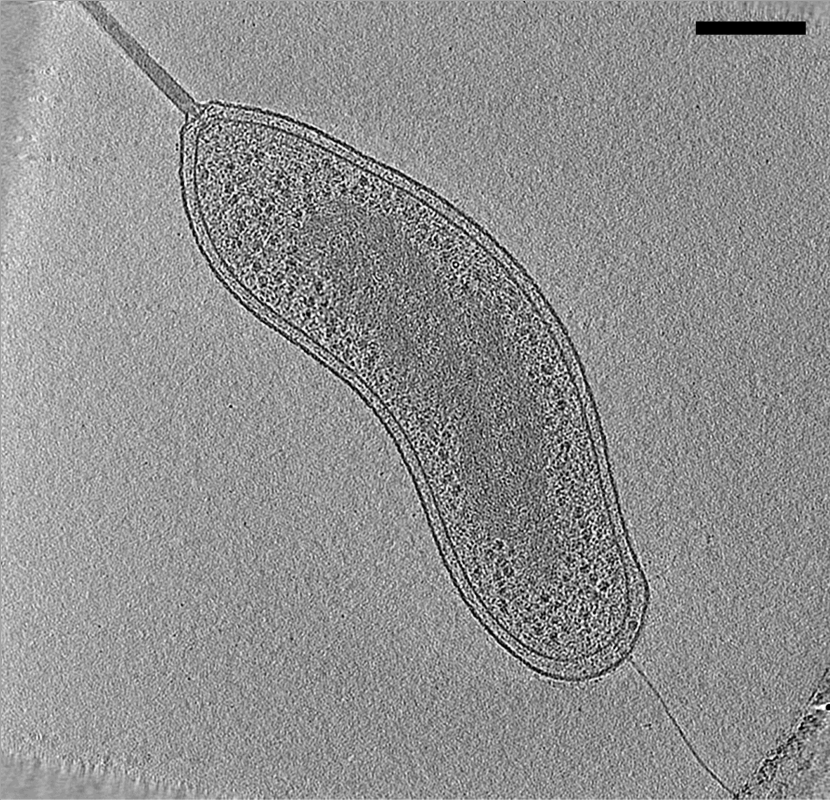
Cryo Electron Tomography
Electron cryotomography (CryoET) is an imaging technique used to produce high-resolution (~1–4 nm) three-dimensional views of samples, often (but not limited to) biological macromolecules and cells. CryoET is a specialized application of transmission electron cryomicroscopy (CryoTEM) in which samples are imaged as they are tilted, resulting in a series of 2D images that can be combined to produce a 3D reconstruction, similar to a CT scan of the human body. In contrast to other electron tomography techniques, samples are imaged under cryogenic conditions (< −150 °C). For cellular material, the structure is immobilized in non-crystalline, vitreous ice, allowing them to be imaged without dehydration or chemical fixation, which would otherwise disrupt or distort biological structures. Description of technique [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Magnetic Resonance Spectroscopy Of Proteins
Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the NIH, and Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated. NMR involves the quantum-mechanical properties of the central core ("nucleus") of the atom. These properties depend on the local molecular environment, and their measurement provides a map of how the atoms are linked chemically, how close they are in space, and how r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Macromolecular Crowding
The phenomenon of macromolecular crowding alters the properties of molecules in a solution when high concentrations of macromolecules such as proteins are present. Such conditions occur routinely in living cells; for instance, the cytosol of ''Escherichia coli'' contains about 300– of macromolecules. Crowding occurs since these high concentrations of macromolecules reduce the volume of solvent available for other molecules in the solution, which has the result of increasing their effective concentrations. Crowding can promote formation of a biomolecular condensate by colloidal phase separation. This crowding effect can make molecules in cells behave in radically different ways than in test-tube assays. Consequently, measurements of the properties of enzymes or processes in metabolism that are made in the laboratory (''in vitro'') in dilute solutions may be different by many orders of magnitude from the true values seen in living cells (''in vivo''). The study of biochemical pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biomolecular Complex
A biomolecule or biological molecule is a loosely used term for molecules present in organisms that are essential to one or more typically biological processes, such as cell division, morphogenesis, or development. Biomolecules include large macromolecules (or polyelectrolytes) such as proteins, carbohydrates, lipids, and nucleic acids, as well as small molecules such as primary metabolites, secondary metabolites and natural products. A more general name for this class of material is biological materials. Biomolecules are an important element of living organisms, those biomolecules are often endogenous, produced within the organism but organisms usually need exogenous biomolecules, for example certain nutrients, to survive. Biology and its subfields of biochemistry and molecular biology study biomolecules and their reactions. Most biomolecules are organic compounds, and just four elements—oxygen, carbon, hydrogen, and nitrogen—make up 96% of the human body's mass. But ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure
Protein structure is the molecular geometry, three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a ''residue'' indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per chemical reaction, reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their Pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations (PDBe, PDBj, RCSB, and BMRB). The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB. The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other databases use protein structures deposited in the PDB. For example, SCOP and CATH classify protein structures, while PDBsum provides a graphic overview of PDB entries using information from other sources, such as Gene ontology. History Two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound. Cofactors can be divided into two types: inorganic ions and complex organic molecules called coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Note that some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a "prosthetic group", which consists of a coenzyme that is tightly (or even covalently) and permanently bound to a prote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding Site
In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity. The binding partner of the macromolecule is often referred to as a ligand. Ligands may include other proteins (resulting in a protein-protein interaction), enzyme substrates, second messengers, hormones, or allosteric modulators. The binding event is often, but not always, accompanied by a conformational change that alters the protein's function. Binding to protein binding sites is most often reversible (transient and non-covalent), but can also be covalent reversible or irreversible. Function Binding of a ligand to a binding site on protein often triggers a change in conformation in the protein and results in altered cellular function. Hence binding site on protein are critical parts of signal transduction pathways. Types of ligands include neurotransmitters, toxins, neuropeptides, and steroid hormones. Binding sit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |