|
DUF3085 RNA Motif
The DUF3085 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF3085 motifs are found in one species in the genus Thauera, as well as various metagenomic sequences obtained from environmental DNA. DUF3085 motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes. The genes they putatively regulate often encode the DUF3085 conserved protein domain or ParB, a protein often used by plasmid A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and ...s. However, because DUF3085 RNAs were not found in any fully sequenced organism, it was not possible to determine if DUF3085 RNAs are consistently located in plasmids. References {{reflist Non-coding RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids (DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst eukaryotes, and the tmRNA in bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, promp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
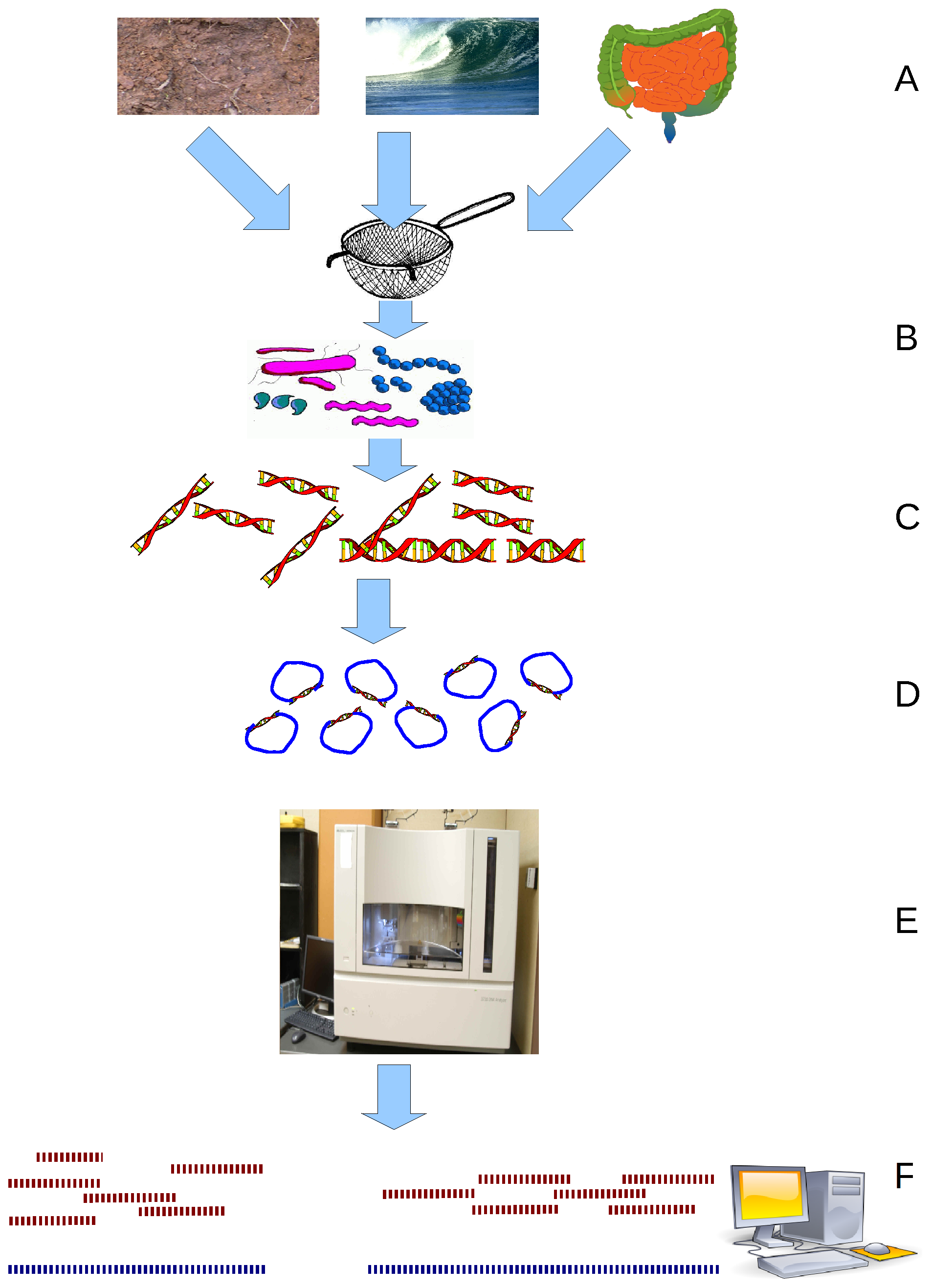
Bioinformatics Discovery Of Non-coding RNAs
Non-coding RNAs have been discovered using both experimental and bioinformatic approaches. Bioinformatic approaches can be divided into three main categories. The first involves homology search, although these techniques are by definition unable to find new classes of ncRNAs. The second category includes algorithms designed to discover specific types of ncRNAs that have similar properties. Finally, some discovery methods are based on very general properties of RNA, and are thus able to discover entirely new kinds of ncRNAs. Discovery by homology search Homology search refers to the process of searching a sequence database for RNAs that are similar to already known RNA sequences. Any algorithm that is designed for homology search of nucleic acid sequences can be used, e.g., BLAST. However, such algorithms typically are not as sensitive or accurate as algorithms specifically designed for RNA. Of particular importance for RNA is its conservation of a secondary structure, which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genus
Genus (; : genera ) is a taxonomic rank above species and below family (taxonomy), family as used in the biological classification of extant taxon, living and fossil organisms as well as Virus classification#ICTV classification, viruses. In binomial nomenclature, the genus name forms the first part of the binomial species name for each species within the genus. :E.g. ''Panthera leo'' (lion) and ''Panthera onca'' (jaguar) are two species within the genus ''Panthera''. ''Panthera'' is a genus within the family Felidae. The composition of a genus is determined by taxonomy (biology), taxonomists. The standards for genus classification are not strictly codified, so different authorities often produce different classifications for genera. There are some general practices used, however, including the idea that a newly defined genus should fulfill these three criteria to be descriptively useful: # monophyly – all descendants of an ancestral taxon are grouped together (i.e. Phylogeneti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thauera
''Thauera'' is a genus of Gram-negative bacteria in the family ''Zoogloeaceae'' of the order ''Rhodocyclales'' of the ''Betaproteobacteria ''Betaproteobacteria'' are a class of Gram-negative bacteria, and one of the six classes of the phylum '' Pseudomonadota'' (synonym Proteobacteria). Metabolism The ''Betaproteobacteria'' comprise over 75 genera and 400 species. Together, they ...''. The genus is named for the German microbiologist Rudolf Thauer. Most species of this genus are motile by flagella and are mostly rod-shaped.Garrity, George M.; Brenner, Don J.; Krieg, Noel R.; Staley, James T. (eds.) (2005). Bergey's Manual of Systematic Bacteriology, Volume Two: The Proteobacteria, Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. New York, New York: Springer. . The species occur in wet soil and polluted freshwater. Species The genus includes the following species: * '' T. aminoaromatica'' * '' T. aromatica'' * '' T. butanivorans'' * '' T. chlorobenzoica'' * ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metagenomic
Metagenomics is the study of all genetic material from all organisms in a particular environment, providing insights into their composition, diversity, and functional potential. Metagenomics has allowed researchers to profile the microbial composition of environmental and clinical samples without the need for time-consuming culture of individual species. Metagenomics has transformed microbial ecology and evolutionary biology by uncovering previously hidden biodiversity and metabolic capabilities. As the cost of DNA sequencing continues to decline, metagenomic studies now routinely profile hundreds to thousands of samples, enabling large-scale exploration of microbial communities and their roles in health and global ecosystems. Metagenomic studies most commonly employ shotgun sequencing though long-read sequencing is being increasingly utilised as technologies advance. The field is also referred to as environmental genomics, ecogenomics, community genomics, or microbiomics a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes. During gene expression (the synthesis of Gene product, RNA or protein from a gene), DNA is first transcription (biology), copied into RNA. RNA can be non-coding RNA, directly functional or be the intermediate protein biosynthesis, template for the synthesis of a protein. The transmission of genes to an organism's offspring, is the basis of the inheritance of phenotypic traits from one generation to the next. These genes make up different DNA sequences, together called a genotype, that is specific to every given individual, within the gene pool of the population (biology), population of a given species. The genotype, along with environmental and developmental factors, ultimately determines the phenotype ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or Disulfide bond, disulfide bridges. Domains often form functional units, such as the calcium-binding EF-hand, EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimera (protein), chimeric ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and archaea; however plasmids are sometimes present in and eukaryotic organisms as well. Plasmids often carry useful genes, such as those involved in antibiotic resistance, virulence, secondary metabolism and bioremediation. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain additional genes for special circumstances. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet by various vendors ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |