|
DIALIGN-TX
DIALIGN-TX is a multiple sequence alignment program written by Amarendran R. Subramanian and is substantial improvement of DIALIGN-T by combining greedy and progressive alignment strategies in a new algorithm. The original DIALIGN-T is a reimplementation of the multiple-alignment program DIALIGN. Due to several algorithmic improvements, it produces significantly better alignments on locally and globally related sequence sets than previous versions of DIALIGN. However, like the original implementation of the program, DIALIGN-T uses a straightforward greedy approach to assemble multiple alignments from local pairwise sequence similarities. Such greedy approaches may be vulnerable to spurious random similarities and can therefore lead to suboptimal results. DIALIGN-TX is a substantial improvement of DIALIGN-T that combines the previous greedy algorithm with a progressive alignment approach. See also * DIALIGN-T *Sequence alignment software *Clustal Clustal is a computer program ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment Software
This list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequence alignment. See structural alignment software for structural alignment of proteins. Database search only *Sequence type: protein or nucleotide Pairwise alignment *Sequence type: protein or nucleotide **Alignment type: local or global Multiple sequence alignment *Sequence type: protein or nucleotide. **Alignment type: local or global Genomics analysis *Sequence type: protein or nucleotide Motif finding *Sequence type: protein or nucleotide Benchmarking Alignment viewers, editors Please see List of alignment visualization software. Short-read sequence alignment See also * List of open source bioinformatics software References {{Reflist Seq Sequence In mathematics, a sequence is an enumerated collection of objects in which repetitions are allowed and order matters. Like a set, it contains members (also called ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clustal
Clustal is a computer program used for multiple sequence alignment in bioinformatics. The software and its algorithms have gone through several iterations, with ClustalΩ (Omega) being the latest version . It is available as standalone software, via a Clustal#External links, web interface, and through a server hosted by the European Bioinformatics Institute. Clustal has been an important bioinformatic software, with two of its academic publications amongst the top 100 papers cited of all time, according to Nature (journal), Nature in 2014. History Version history * Clustal: The original software for multiple sequence alignments, created by Desmond G. Higgins, Des Higgins in 1988, was based on deriving a guide tree from pairwise sequences of Amino acid, amino acids or Nucleotide, nucleotides. * ClustalV: The second generation of Clustal, released in 1992. It introduced the ability to create new alignments from existing alignments in a process known as phylogenetic tree re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
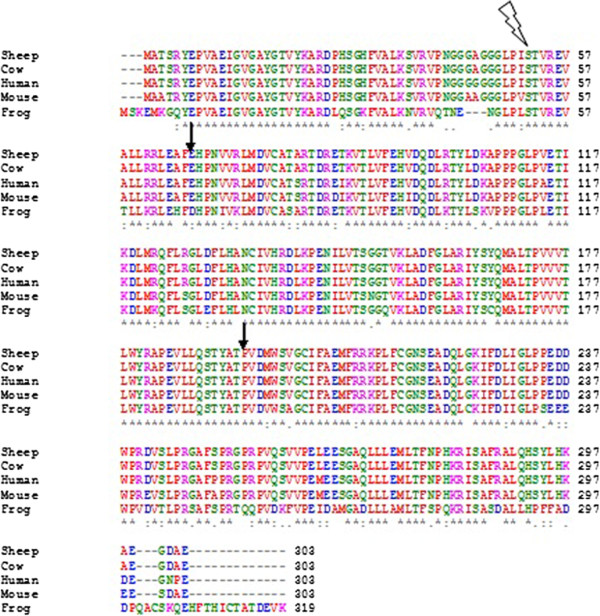
Multiple Sequence Alignment
Multiple sequence alignment (MSA) is the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. These alignments are used to infer evolutionary relationships via phylogenetic analysis and can highlight homologous features between sequences. Alignments highlight mutation events such as point mutations (single amino acid or nucleotide changes), insertion mutations and deletion mutations, and alignments are used to assess sequence conservation and infer the presence and activity of protein domains, tertiary structures, secondary structures, and individual amino acids or nucleotides. Multiple sequence alignments require more sophisticated methodologies than pairwise alignments, as they are more computationally complex. Most multiple sequence alignment programs use heuristic methods rather than global optimization because identifying the optimal alignment between more than a few sequences of moderate length is prohibiti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |