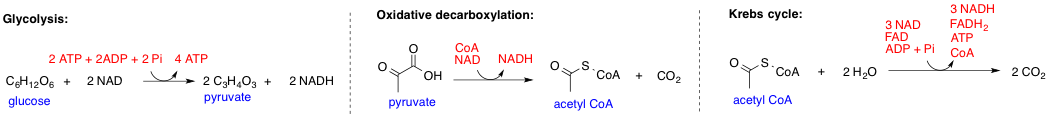|
Cpdb Network
The ConsensusPathDB is a molecular functional interaction database, integrating information on protein interactions, genetic interactions signaling, metabolism, gene regulation, and drug-target interactions in humans. ConsensusPathDB currently (release 30) includes such interactions from 32 databases. ConsensusPathDB is freely available for academic use under http://ConsensusPathDB.org. Integrated Databases * Reactome (metabolic and signaling pathways) * KEGG (metabolic pathways only have been integrated in ConsensusPathDB) * HumanCyc (metabolic pathways) * PID - Pathway Interaction Database (signaling pathways) * BioCarta (signaling pathways) * Netpath (signaling pathways) * IntAct (protein interactions) * DIP (protein interactions) * MINT (protein interactions) * HPRD (protein interactions) * BioGRID (protein interactions) * SPIKE (protein interactions, signaling reactions) * WikiPathways (metabolic and signaling pathways) * and many more. Functionalities The ConsensusPathDB is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Database
In computing, a database is an organized collection of data stored and accessed electronically. Small databases can be stored on a file system, while large databases are hosted on computer clusters or cloud storage. The design of databases spans formal techniques and practical considerations, including data modeling, efficient data representation and storage, query languages, security and privacy of sensitive data, and distributed computing issues, including supporting concurrent access and fault tolerance. A database management system (DBMS) is the software that interacts with end users, applications, and the database itself to capture and analyze the data. The DBMS software additionally encompasses the core facilities provided to administer the database. The sum total of the database, the DBMS and the associated applications can be referred to as a database system. Often the term "database" is also used loosely to refer to any of the DBMS, the database system or an appli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Protein Reference Database
The Human Protein Reference Database (HPRD) is a protein database accessible through the Internet. It is closely associated with the premier Indian Non-Profit research organisation Institute of Bioinformatics (IOB), Bangalore. This database is a collaborative output of IOB and the Pandey Lab of Johns Hopkins University. Overview The HPRD is a result of an international collaborative effort between the Institute of Bioinformatics in Bangalore, India and the Pandey lab at Johns Hopkins University in Baltimore, USA. HPRD contains manually curated scientific information pertaining to the biology of most human proteins. Information regarding proteins involved in human diseases is annotated and linked to Online Mendelian Inheritance in Man (OMIM) database. The National Center for Biotechnology Information provides link to HPRD through its human protein databases (e.g. Entrez Gene, RefSeq protein pertaining to genes and proteins. This resource depicts information on human protein functi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
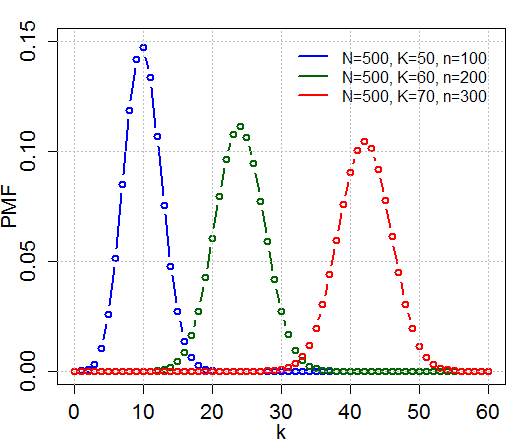
Hypergeometric Distribution
In probability theory and statistics, the hypergeometric distribution is a discrete probability distribution that describes the probability of k successes (random draws for which the object drawn has a specified feature) in n draws, ''without'' replacement, from a finite population of size N that contains exactly K objects with that feature, wherein each draw is either a success or a failure. In contrast, the binomial distribution describes the probability of k successes in n draws ''with'' replacement. Definitions Probability mass function The following conditions characterize the hypergeometric distribution: * The result of each draw (the elements of the population being sampled) can be classified into one of two mutually exclusive categories (e.g. Pass/Fail or Employed/Unemployed). * The probability of a success changes on each draw, as each draw decreases the population ('' sampling without replacement'' from a finite population). A random variable X follows the hype ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P-value
In null-hypothesis significance testing, the ''p''-value is the probability of obtaining test results at least as extreme as the result actually observed, under the assumption that the null hypothesis is correct. A very small ''p''-value means that such an extreme observed outcome would be very unlikely under the null hypothesis. Reporting ''p''-values of statistical tests is common practice in academic publications of many quantitative fields. Since the precise meaning of ''p''-value is hard to grasp, misuse is widespread and has been a major topic in metascience. Basic concepts In statistics, every conjecture concerning the unknown probability distribution of a collection of random variables representing the observed data X in some study is called a ''statistical hypothesis''. If we state one hypothesis only and the aim of the statistical test is to see whether this hypothesis is tenable, but not to investigate other specific hypotheses, then such a test is called a null ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biochemical Pathway
In biochemistry, a metabolic pathway is a linked series of chemical reactions occurring within a cell. The reactants, products, and intermediates of an enzymatic reaction are known as metabolites, which are modified by a sequence of chemical reactions catalyzed by enzymes. In most cases of a metabolic pathway, the product of one enzyme acts as the substrate for the next. However, side products are considered waste and removed from the cell. These enzymes often require dietary minerals, vitamins, and other cofactors to function. Different metabolic pathways function based on the position within a eukaryotic cell and the significance of the pathway in the given compartment of the cell. For instance, the, electron transport chain, and oxidative phosphorylation all take place in the mitochondrial membrane. In contrast, glycolysis, pentose phosphate pathway, and fatty acid biosynthesis all occur in the cytosol of a cell. There are two types of metabolic pathways that are characterize ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
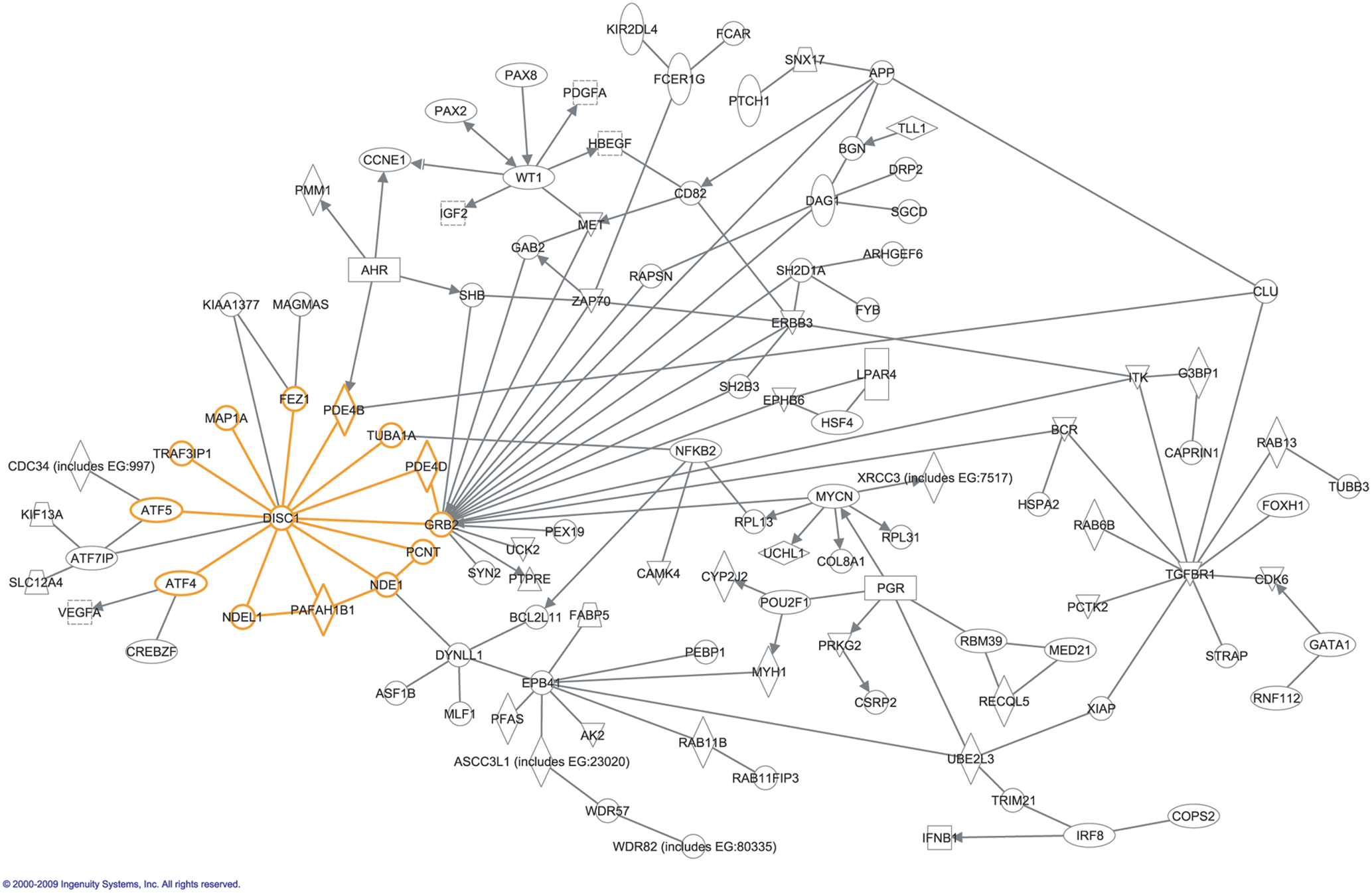
Interaction Network
In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. The term specifically refers to physical interactions among molecules (such as those among proteins, also known as protein–protein interactions, PPIs; or between small molecules and proteins) but can also describe sets of indirect interactions among genes ( genetic interactions). The word "interactome" was originally coined in 1999 by a group of French scientists headed by Bernard Jacq. Mathematically, interactomes are generally displayed as graphs. Though interactomes may be described as biological networks, they should not be confused with other networks such as neural networks or food webs. Molecular interaction networks Molecular interactions can occur between molecules belonging to different biochemical families (proteins, nucleic acids, lipids, carbohydrates, etc.) and also within a given family. Whenever such molecules are connected by physical interactions, they form molec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cpdb Network
The ConsensusPathDB is a molecular functional interaction database, integrating information on protein interactions, genetic interactions signaling, metabolism, gene regulation, and drug-target interactions in humans. ConsensusPathDB currently (release 30) includes such interactions from 32 databases. ConsensusPathDB is freely available for academic use under http://ConsensusPathDB.org. Integrated Databases * Reactome (metabolic and signaling pathways) * KEGG (metabolic pathways only have been integrated in ConsensusPathDB) * HumanCyc (metabolic pathways) * PID - Pathway Interaction Database (signaling pathways) * BioCarta (signaling pathways) * Netpath (signaling pathways) * IntAct (protein interactions) * DIP (protein interactions) * MINT (protein interactions) * HPRD (protein interactions) * BioGRID (protein interactions) * SPIKE (protein interactions, signaling reactions) * WikiPathways (metabolic and signaling pathways) * and many more. Functionalities The ConsensusPathDB is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UniProt
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, United States. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metabolite
In biochemistry, a metabolite is an intermediate or end product of metabolism. The term is usually used for small molecules. Metabolites have various functions, including fuel, structure, signaling, stimulatory and inhibitory effects on enzymes, catalytic activity of their own (usually as a cofactor to an enzyme), defense, and interactions with other organisms (e.g. pigments, odorants, and pheromones). A primary metabolite is directly involved in normal "growth", development, and reproduction. Ethylene exemplifies a primary metabolite produced large-scale by industrial microbiology. A secondary metabolite is not directly involved in those processes, but usually has an important ecological function. Examples include antibiotics and pigments such as resins and terpenes etc. Some antibiotics use primary metabolites as precursors, such as actinomycin, which is created from the primary metabolite tryptophan. Some sugars are metabolites, such as fructose or glucose, which ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid resid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Physical Entity
In common usage and classical mechanics, a physical object or physical body (or simply an object or body) is a collection of matter within a defined contiguous boundary in three-dimensional space. The boundary must be defined and identified by the properties of the material. The boundary may change over time. The boundary is usually the visible or tangible surface of the object. The matter in the object is constrained (to a greater or lesser degree) to move as one object. The boundary may move in space relative to other objects that it is not attached to (through translation and rotation). An object's boundary may also deform and change over time in other ways. Also in common usage, an object is not constrained to consist of the same collection of matter. Atoms or parts of an object may change over time. An object usually meant to be defined by the simplest representation of the boundary consistent with the observations. However the laws of physics only apply directly to objects ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Web Interface
In the industrial design field of human–computer interaction, a user interface (UI) is the space where interactions between humans and machines occur. The goal of this interaction is to allow effective operation and control of the machine from the human end, while the machine simultaneously feeds back information that aids the operators' decision-making process. Examples of this broad concept of user interfaces include the interactive aspects of computer operating systems, hand tools, heavy machinery operator controls and process controls. The design considerations applicable when creating user interfaces are related to, or involve such disciplines as, ergonomics and psychology. Generally, the goal of user interface design is to produce a user interface that makes it easy, efficient, and enjoyable (user-friendly) to operate a machine in the way which produces the desired result (i.e. maximum usability). This generally means that the operator needs to provide minimal input to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |