|
Co-activator
A coactivator is a type of transcriptional coregulator that binds to an activator (a transcription factor) to increase the rate of transcription of a gene or set of genes. The activator contains a DNA binding domain that binds either to a DNA promoter site or a specific DNA regulatory sequence called an enhancer. Binding of the activator-coactivator complex increases the speed of transcription by recruiting general transcription machinery to the promoter, therefore increasing gene expression. The use of activators and coactivators allows for highly specific expression of certain genes depending on cell type and developmental stage. Some coactivators also have histone acetyltransferase (HAT) activity. HATs form large multiprotein complexes that weaken the association of histones to DNA by acetylating the N-terminal histone tail. This provides more space for the transcription machinery to bind to the promoter, therefore increasing gene expression. Activators are found in all ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EP300
Histone acetyltransferase p300 also known as p300 HAT or E1A-associated protein p300 (where E1A = adenovirus early region 1A) also known as EP300 or p300 is an enzyme that, in humans, is encoded by the ''EP300'' gene. It functions as histone acetyltransferase that regulates transcription of genes via chromatin remodeling by allowing histone proteins to wrap DNA less tightly. This enzyme plays an essential role in regulating cell growth and division, prompting cells to mature and assume specialized functions (differentiate), and preventing the growth of cancerous tumors. The p300 protein appears to be critical for normal development before and after birth. The EP300 gene is located on the long (q) arm of the human chromosome 22 at position 13.2. This gene encodes the adenovirus E1A-associated cellular p300 transcriptional co-activator protein. EP300 is closely related to another gene, CREB binding protein, which is found on human chromosome 16. Function p300 HAT funct ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Acetyltransferase
Histone acetyltransferases (HATs) are enzymes that acetylation, acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-N-acetyllysine, ε-''N''-acetyllysine. DNA is wrapped around histones, and, by transferring an acetyl group to the histones, genes can be turned on and off. In general, histone acetylation increases gene expression. In general, histone acetylation is linked to DNA transcription, transcriptional activation and associated with euchromatin. Euchromatin, which is less densely compact, allows transcription factors to bind more easily to regulatory sites on DNA, causing transcriptional activation. When it was first discovered, it was thought that acetylation of lysine neutralizes the positive electric charge, charge normally present, thus reducing affinity between histone and (negatively charged) DNA, which renders DNA more accessible to transcription factors. Research has emerged, since, to show that lys ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
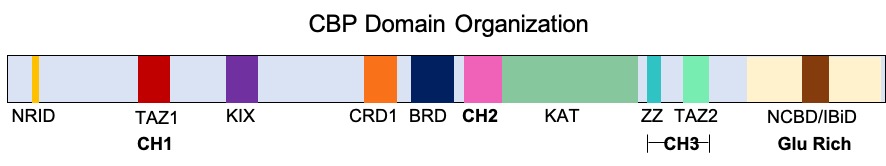
CREB-binding Protein
CREB-binding protein, also known as CREBBP or CBP or KAT3A, (where CREB is cAMP response element-binding protein) is a coactivator encoded by the ''CREBBP'' gene in humans, located on chromosome 16p13.3. CBP has intrinsic acetyltransferase functions; it is able to add acetyl groups to both transcription factors as well as histone lysines, the latter of which has been shown to alter chromatin structure making genes more accessible for transcription. This relatively unique acetyltransferase activity is also seen in another transcription enzyme, EP300 (p300). Together, they are known as the p300-CBP coactivator family and are known to associate with more than 16,000 genes in humans; however, while these proteins share many structural features, emerging evidence suggests that these two co-activators may promote transcription of genes with different biological functions. For example, CBP alone has been implicated in a wide variety of pathophysiologies including colorectal cancer as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Overview For transcription to take place, the enzyme that synthesizes RNA, known as RNA polymerase, must attach to the DNA near a gene. Promoters contain specific DNA sequences such as response elements that provide a secure initial binding site for RNA polymerase and for proteins called transcription factors that recruit RNA polymerase. These transcription factor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enhancer (genetics)
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are ''cis''-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes. Active enhancers typically get transcribed as enhancer or regulatory non-coding RNA, whose expression levels correlate with mRNA levels of target genes. The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983. This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive. Lately, enhancers have been shown to be in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transcribed into RNA molecules called non-coding RNAs (ncRNAs). Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a Complementarity (molecular biology), complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, Antiparallel (biochemistry), antiparallel RNA strand called a primary transcript. In virology, the term transcription is used when referring to mRNA synthesis from a viral RNA molecule. The genome of many Orthornavirae, RNA viruses is composed of Sense (molecular biology), negative-sense RNA which acts as a template for positive sense viral messenger RNA - a necessary step in the synthesis of viral proteins needed for viral replication. This process ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulatory Sequence
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses. Description In DNA, regulation of gene expression normally happens at the level of RNA biosynthesis ( transcription). It is accomplished through the sequence-specific binding of proteins ( transcription factors) that activate or inhibit transcription. Transcription factors may act as activators, repressors, or both. Repressors often act by preventing RNA polymerase from forming a productive complex with the transcriptional initiation region ( promoter), while activators facilitate formation of a productive complex. Furthermore, DNA motifs have been shown to be predictive of epigenomic modifications, suggesting that transcription factors play a role in regulating the epigenome. In RNA, regulation may occur at the level of protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, and ultimately affect a phenotype. These products are often proteins, but in non-protein-coding genes such as Transfer RNA, transfer RNA (tRNA) and Small nuclear RNA, small nuclear RNA (snRNA), the product is a functional List of RNAs, non-coding RNA. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and viruses—to generate the macromolecule, macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, ''i.e.'' observable trait. The genetic information stored in DNA represents the genotype, whereas the phenotype results from the "interpretation" of that informati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CARM1
CARM1 (coactivator-associated arginine methyltransferase 1), also known as PRMT4 (protein arginine N-methyltransferase 4), is an enzyme () encoded by the gene found in human beings, as well as many other mammals. It has a polypeptide (L) chain type that is 348 residues long, and is made up of alpha helices and beta sheets. Its main function includes catalyzing the transfer of a methyl group from S-Adenosyl methionine to the side chain nitrogens of arginine residues within proteins to form methylated arginine derivatives and S-Adenosyl-L-homocysteine. CARM1 is a secondary coactivator through its association with p160 family ( SRC-1, GRIP1, AIB) of coactivators. It is responsible for moving cells toward the inner cell mass in developing blastocysts. Clinical significance CARM1 plays an important role in androgen receptors and may play a role in prostate cancer progression. CARM1 exerts both oncogenic and tumor-suppressive functions. In breast cancer, CARM1 methylates chromatin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Corepressor
In genetics and molecular biology, a corepressor is a molecule that represses the expression of genes. In prokaryotes A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ..., corepressors are Small molecule, small molecules whereas in Eukaryote, eukaryotes, corepressors are Protein, proteins. A corepressor does not directly bind to DNA, but instead indirectly regulates gene expression by binding to Repressor, repressors. A corepressor Downregulation and upregulation, downregulates (or represses) the expression of genes by binding to and activating a repressor transcription factor. The repressor in turn binds to a gene's Operator (biology), operator sequence (segment of DNA to which a transcription factor binds to regulate gene expression), thereby blocking transcription of that gene. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Type Ii Nuclear Receptor Action
Type may refer to: Science and technology Computing * Typing, producing text via a keyboard, typewriter, etc. * Data type, collection of values used for computations. * File type * TYPE (DOS command), a command to display contents of a file. * Type (Unix), a command in POSIX shells that gives information about commands. * Type safety, the extent to which a programming language discourages or prevents type errors. * Type system, defines a programming language's response to data types. Mathematics * Type (model theory) * Type theory, basis for the study of type systems * Arity or type, the number of operands a function takes * Type, any proposition or set in the intuitionistic type theory * Type, of an entire function#Order and type, entire function ** Exponential type Biology * Type (biology), which fixes a scientific name to a taxon * Dog type, categorization by use or function of domestic dogs Lettering * Type is a design concept for lettering used in typography which helped b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |